Methods to selectively deplete or purify Hemoglobin from Dried Blood Spots (DBS)
Background
Dried blood spot (DBS) analysis offers the advantage of collecting small blood volumes without the need for phlebotomy. DBS cards can placed in an envelope for simple and cost effective transport, as they do not require temperature regulation. This negates the often cumbersome and expensive processes associated with transport of liquid biological samples. Once the DBS samples are received, the size and properties of the DBS samples make storage relatively easy as minimal space is required, and they usually can be stored at room temperature. Mass spectrometry is now the most common technique reported in the literature for dried blood spot analysis. While DBS mass spectrometry has largely been for newborn screening of metabolic disorders, the role of DBS analysis by MS now encompasses translational research and clinical diagnostic analytes in the areas of therapeutic drug monitoring, pharmakinetics, toxicokinetics, endocrinology, metabolism, and other areas of bio-analysis1. In addition, specific protein(s) and proteomic profiles are now under investigation from DBS samples. For this, simple methods to either deplete Hemoglobin or as in the case of Hemoglobinopathies, isolate Hemoglobin, are often required.
Challenge Due to its spectral properties, Hemoglobin can often pose analytical challenges when either the analyte of interest or the reporting measurement spectrally overlap. This can often be the case for bilirubin, enzyme and ELISA analyses. In addition to its spectral properties, Hemoglobin is the highest abundance protein in whole blood, so any proteomic investigations derived from whole blood, such as DBS, can be severely compromised by its high abundance. On the other side of the spectrum is the case of Hemoglobinopathy and thalassemia, inherited disorders related to the structural variation of hemoglobin (Hb) in red blood cells. Mass spectrometry (MS)-based assays for analysis of hemoglobinopathies have been developed to overcome the limitations of traditional methods. MS can be used to detect the mass/charge ratio (m/z) and response intensity of Hb-specific peptides to achieve qualitative and quantitative detection of Hb subunits. In one report, Hb components were extracted from a dried blood card disc punched (3.2 mm diameter) from the DBS samples and digested by trypsin, to measure a series of Hb- proteo-specific peptides by HPLC–HRMS analysis1. Early detection by prenatal diagnosis or pre-symptomatic screening may reduce the incidence and harm of these diseases. Therefore, clinical applications of MS for first-line screening of hemoglobinopathies are anticipated. For this, the isolation of Hemoglobin may assist in the analysis, as shown in one of the following featured reports.
Solution As one of the key BSG Advantages, BSG offers a choice of two strategies for proteome enrichment, to get the best results: either selectively binding high abundance proteins (the “Bind” products), or choosing not to bind (the “Void” products) thereby enriching the underlying low-abundance sub-proteome. For Hemoglobin depletion, we offer: >the “Bind” products HemogloBind™ & NuGel™ HemogloBind™, derived from a class of solid-phase elastomeric poly-electrolytyes that selectively binds hemoglobin. Without the use of antibodies or biologically derived ligands, HemogloBind™ provides a high degree of selectivity towards Hemoglobin and does not cross react with most common serum components, making it an excellent tool in numerous applications. HemogloBind™ is available in a suspension format, or the NuGel™ porous silica dry-bead format. And, >the “Void” product HemoVoid™, is designed to remove hemoglobin from erythrocyte lysates in a simple and efficient manner, through a negative selection strategy, so that the Hemoglobin voids out through the beads, and the vast majority of the remaining proteome binds to the beads. For subsequent analysis, the bead-bound enriched proteome can either be eluted, or for LC-MS/MS, digested on-bead. This latter process is called Bead-assisted Sample Prep or BASP™.
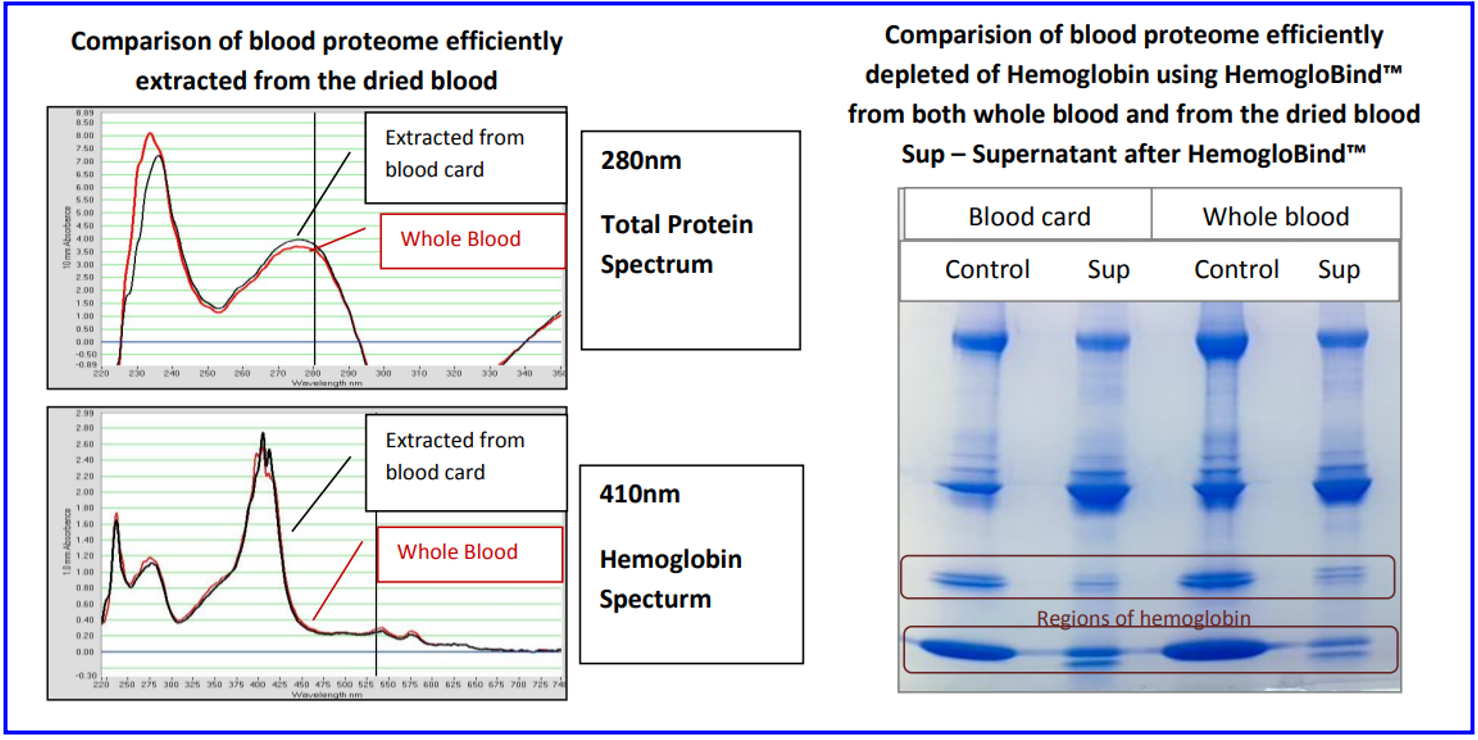
Outcomes The efficiency of HemogloBind™ and HemoVoid™ for whole blood analytes and dried blood cards are noteworthy in many research articles, including:
From: Performance Characteristics of HemogloBind™ on Whole Blood and DBS
To purify Hemoglobin, in a nano LC−MALDI MS/MS measurement of single amino acid mutations in Hb variants. 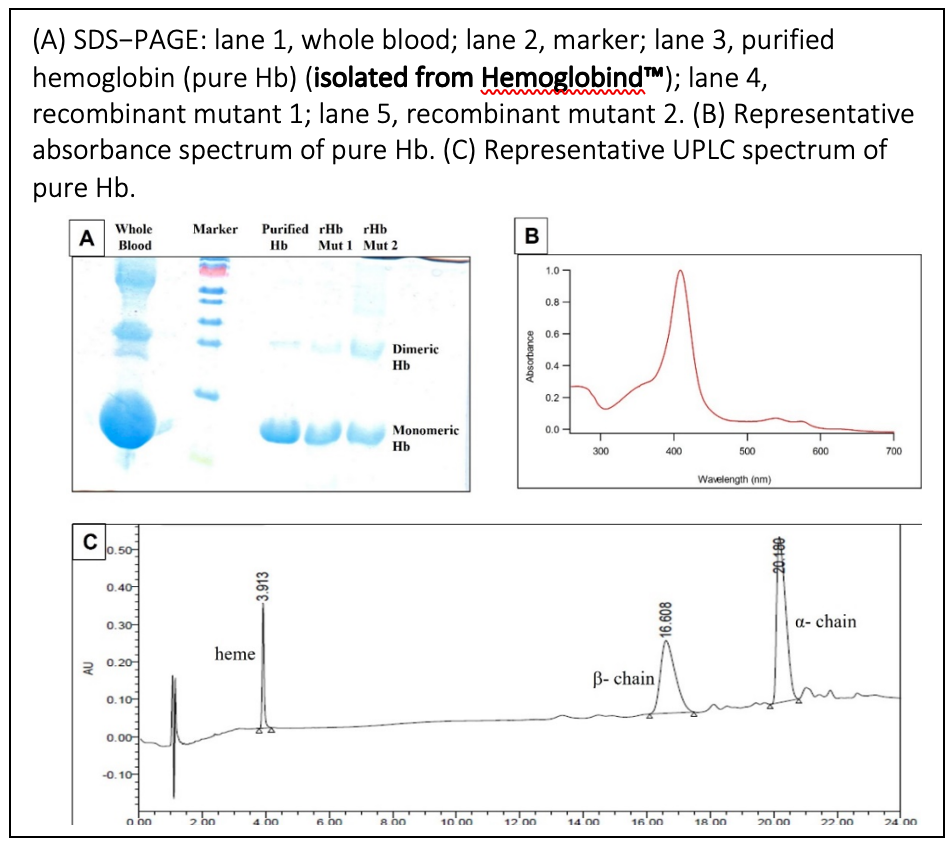
Hemoglobin (Hb) variants arise due to point mutations in globin chains and their pathological treatments rely heavily on the identification of the nature and location of the mutation in the globin chains. Because there is high sequence homology between normal Hb and Hb variant chains, identification of variants by mass spectrometry is very difficult and requires the full sequence coverage of α− and β−chains. Therefore, the present study aims to develop and optimize a specific method of sample processing that could lead to improved sequence coverage and analysis of Hb variants by nano LC−MALDI MS/MS. The article states “Pure Hb was isolated from hemolysate by using Hemoglobind … The Hb bound to the matrix was eluted by increasing the pH of the buffer using 100 mM Tris borate, pH 9. This allows for easy, one−step isolation of Hb from hemolysate.” “The use of Hemoglobind reduced the time to obtain pure Hb in an easy single–step procedure. Pure Hb protein is required specifically to optimize and standardize methods for diagnostics. … The desorbed Hb was compatible with LC–MS, and other proteomics studies, as we verified, did not show any change in its intact mass either. This one−step affinity purification gave us the utmost purified Hb.” The article concludes that a sensitive, robust, and reproducible method was developed to identify/ characterize single substitution mutations in the Hb disorder variants. It simply used pure Hb isolated from blood in a single step by using the Hemoglobind matrix, followed by TFE based sample processing and digestion by trypsin, followed by nano−LC separation. The separated peptides were spotted onto MALDI plates upon elution from a nano−LC column and subjected to MALDI MS and MS/MS analysis. The method highlights a non−targeted approach for the identification of Hb variants and its reproducibility to yield 100% sequence coverage for the β−chain with precise identification of all the mutations. It has the potential to become a regular screening/diagnosis tool for the identification of single amino acid mutations in Hb variants.
From: Dasauni, Pushpanjali, et al. "Optimization and Identification of Single Mutation in Hemoglobin Variants with 2, 2, 2 Trifluoroethanol Modified Digestion Method and Nano− LC Coupled MALDI MS/MS." Molecules 27.19 (2022): 6357.
For Whole Blood Analytes “For protein isolation, whole blood aliquots were lysed with water and haemoglobin was removed using HemogloBind™ (Biotech Support Group) according to manufacturer’s instructions…”. From: Chalásová, Katarína, et al. "Transketolase Activity but not Thiamine Membrane Transport Change in Response to Hyperglycaemia and Kidney Dysfunction." Experimental and Clinical Endocrinology & Diabetes (2017). https://www.thieme-connect.com/products/ejournals/abstract/10.1055/s-0043-115009 For Cholinesterase Analysis from whole blood lysates: “Briefly, whole blood samples were treated with HemogloBind™ which interferes with the ChE activity assay due to spectral overlap.” From: McGarry, Kevin G., et al. "Evaluation of HemogloBind™ treatment for preparation of samples for cholinesterase analysis." (2013). Advances in Bioscience and Biotechnology, 2013, 4, 1020-1023; and Brittain, Matthew K., Kevin G. McGarry, Robert A. Moyer, Michael C. Babin, David A. Jett, Gennady E. Platoff, and David T. Yeung. "Efficacy of Recommended Prehospital Human Equivalent Doses of Atropine and Pralidoxime Against the Toxic Effects of Carbamate Poisoning in the Hartley Guinea Pig." International journal of toxicology (2016): 1091581816638086.
For Dried Blood Spots After comparison of methods, “…the use of HemoVoid™ resulted in the cleanest samples, closely followed by HemogloBind™ (hemoglobin removal and capture reagent), schematic overview…”
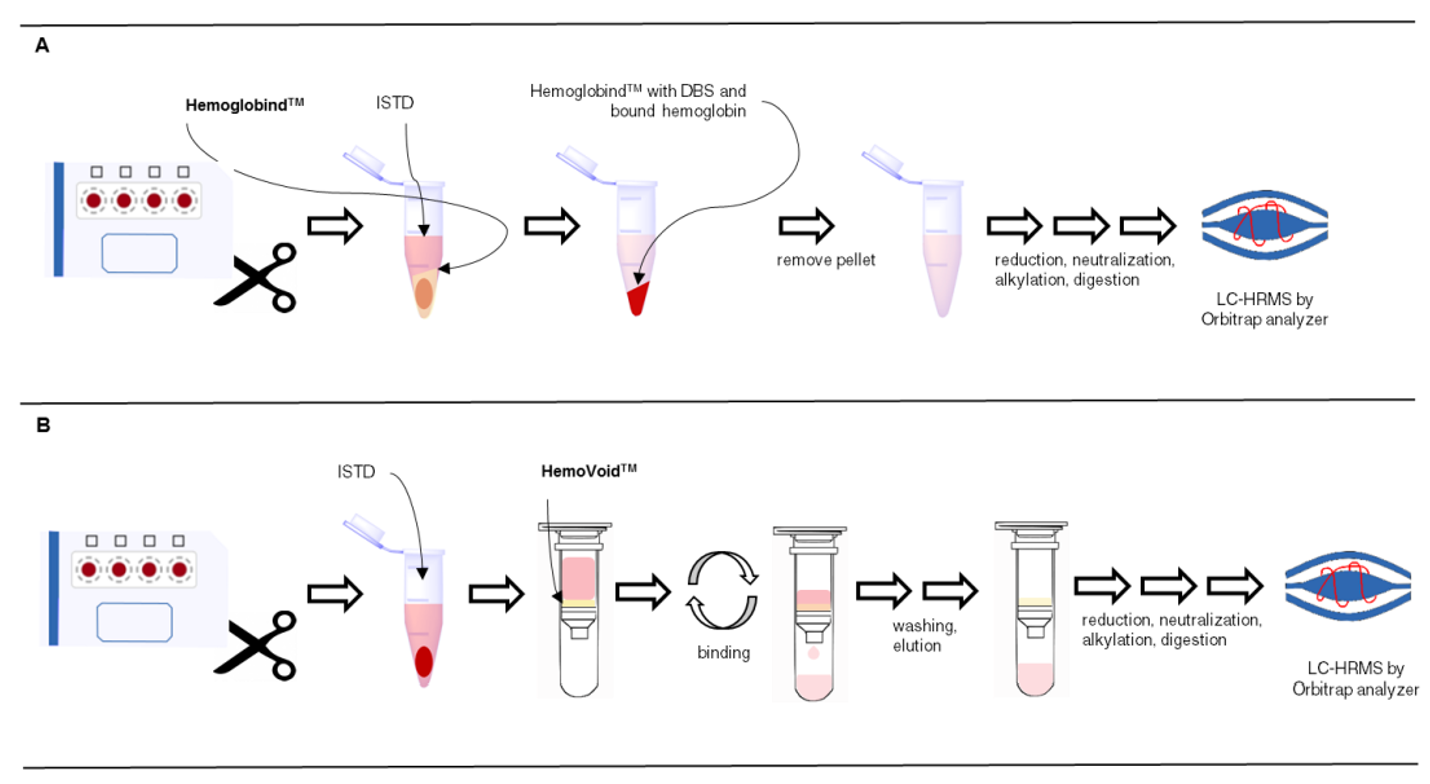 From: Lange, Tobias, et al. "Development of two complementary LC–HRMS methods for analyzing sotatercept in dried blood spots for doping controls." Bioanalysis 11.10 (2019): 923-940.
For Dried Blood Spots "After hemoglobin depletion (using HemogloBind™), panel coverage in VAMS and DBS approaches that observed for traditional plasma samples.". From: Michelle R. Robinson, Lei Guo; Raymond J. Gonzalez; Kara M. Pearson; Kevin P. Bateman; Daniel S. Spellman. Differentiating Modes of Drug Induced Liver Injury Using Parallel Reaction Monitoring LC-MS. ASMS Conference 2017 poster report. "…Hb can be removed using a commercial product, HemogloBind™, which can isolate and remove up to 90% of blood Hb." From: Hakuna, Lovemore, et al. "A simple assay for glutathione in whole blood."Analyst (2015). (http://pubs.rsc.org/en/content/articlelanding/2015/an/c5an00345h)
“Cards were dried, protein extracted and Hb-depleted using HemoVoid™” From: Dried blood spots for detection of autologous blood doping with targeted LC-MS/MS
The HemogloBind™ Blood Card Kit, includes the Protein Extraction Buffer, the HemogloBind™ suspension and spin-filters. 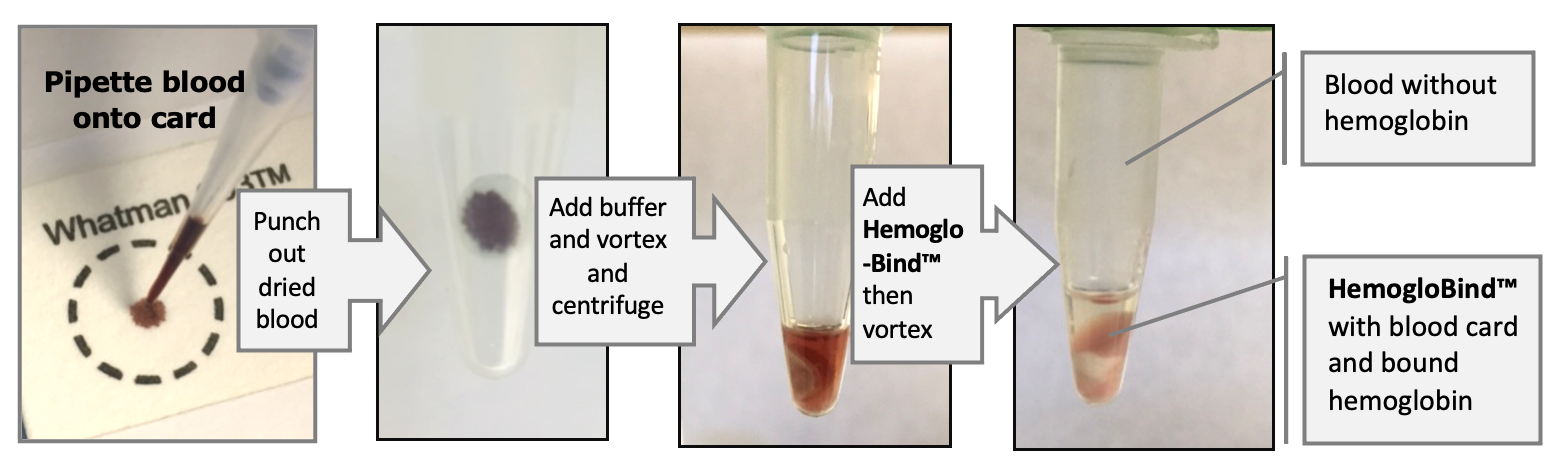
HemoVoid™ - Blood Card Reagent Kit includes all necessary bind, wash, elute buffers, HemoVoid™ beads and spin filters Related Separations, Enrichment/Depletion & Sample Prep - All Product Categories (https://www.biotechsupportgroup.com/Products-a-z_a/258.htm) Albumin & IgG Removal (https://www.biotechsupportgroup.com/Articles.asp?ID=451) Lipid Removal and Clarification (https://www.biotechsupportgroup.com/Articles.asp?ID=456) Hemoglobin Removal (https://www.biotechsupportgroup.com/Articles.asp?ID=452) Sample Prep – Liquid Biopsy (https://www.biotechsupportgroup.com/Articles.asp?ID=457) Sample Prep – Glyco, Virus, Kinase, Aqueous Protein Crash/Metabolomics (https://www.biotechsupportgroup.com/Articles.asp?ID=453) Sample Prep – Mass Spectrometry (https://www.biotechsupportgroup.com/Articles.asp?ID=432) Sample Prep – Genomics (https://www.biotechsupportgroup.com/Articles.asp?ID=455)
References - 1. Zakaria R, Allen KJ, Koplin JJ, Roche P, Greaves RF. Advantages and Challenges of Dried Blood Spot Analysis by Mass Spectrometry Across the Total Testing Process. EJIFCC. 2016 Dec 1;27(4):288-317. PMID: 28149263; PMCID: PMC5282914.
- 2. Li, Ziwei, Chen, Deling, Shu, Yan, Yang, Jing, Zhang, Juan, Ming wang,, Wan, Kexing, Zhou, Yinpin, He, Xiaoyan, Zou, Lin and Yu, Chaowen. "A reliable and high throughput HPLC–HRMS method for the rapid screening of β-thalassemia and hemoglobinopathy in dried blood spots" Clinical Chemistry and Laboratory Medicine (CCLM), 2023. https://doi.org/10.1515/cclm-2022-0706
|