Functional Proteomics 2DE
Using the ArrayBridge PEP separations platform
The unique ArrayBridge PEP platform characterizes function on a proteome scale.
The PEP-based functional proteomics services can be used for:
- tissue specific functional protein annotations
- drug target identification and validation
- drug safety studies and biomarker discovery
- Multi-functional proteome annotation
 The unique ArrayBridge PEP platform characterizes function on a proteome scale.
The unique ArrayBridge PEP platform characterizes function on a proteome scale.
"We have used several of the BSG enrichment product upstream to our PEP platform, and they all help to produce more enzyme features. This is important to our work as the constitution of features is what drives biomarker discovery."
- Xing Wang, Ph.D., President, Array Bridge
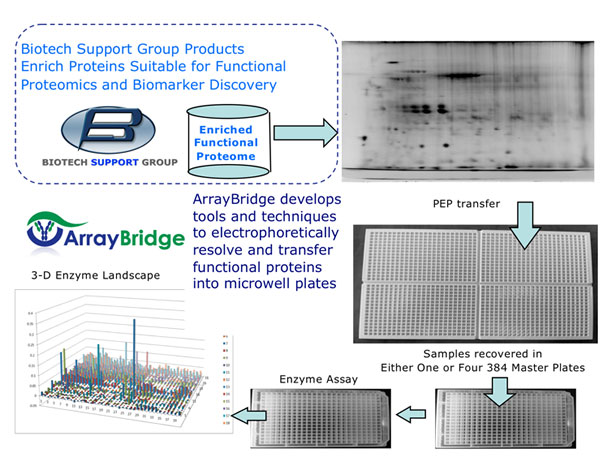
Most efforts in proteomics seek to identify and sequence annotate a proteome by LC-MS/MS analyses of peptides derived through proteolytic processing. However, little attention has been paid to functional annotation. Yet functional annotation is crucial as the landscape of protein conformations is highly variable, each conformation contributing to its own functional activity. Such functional annotation is not strictly proportional to protein level abundances, but rather comes from contributions of structure, interaction and abundances. As a result, quantitative sequence annotation alone cannot capture vital functional information, so new strategies are necessary. The PEP technology, developed by Array Bridge, uses a modified Two-dimensional Gel Electrophoresis to separate the proteome, without substantially compromising function. The isolated proteins are then electro-eluted from the PEP plate, and enzyme activities are measured systematically.
The PEP technology is an open platform adaptable to any measurable protein function. It serves as an efficient method for high sensitivity proteome pattern profiling, quantitative and systematic top-down analysis of isolated protein function, and purity suitable for Mass Spectrometry protein identification. Furthermore, in different tissues, or under stress or disease conditions, protein sequences that are normally associated with one function, may perform alternative functions - that is what we call the multi-functional proteome. This multi-functional proteome can now be monitored, cataloged and annotated by this PEP-based functional proteomic strategy. Using a top-down approach, we can start with any measurable protein function, resolve functionally active proteins into microwells, and from there, sequence and structurally annotate the subset proteome that presents that function.
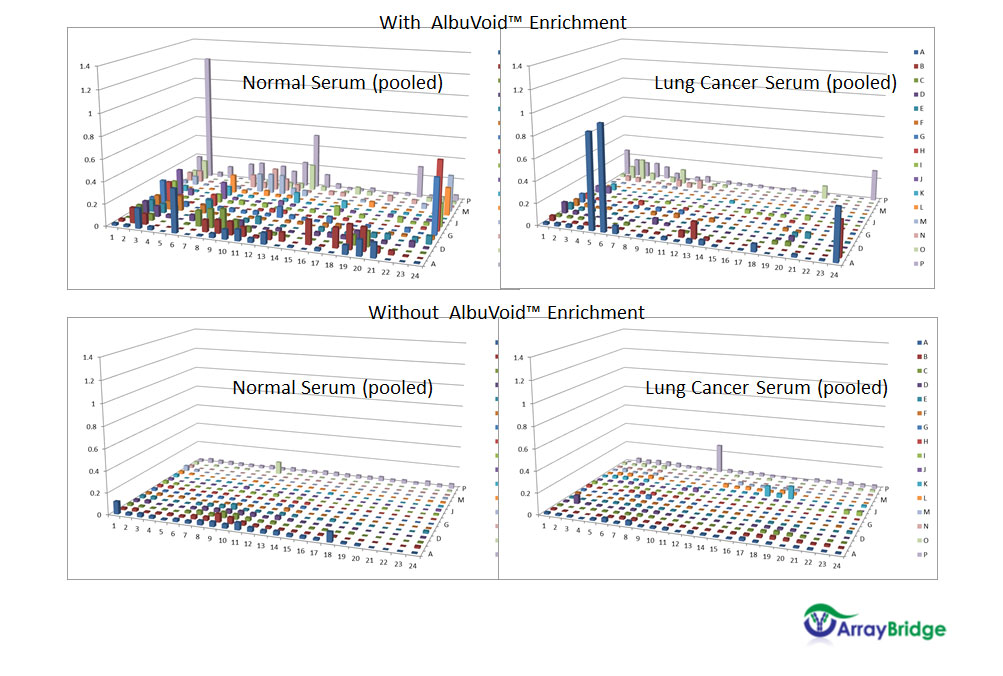
Hexokinase Activity
Normal and Lung Cancer patient Serum With & Without Albuvoid Enrichment
Procedure:
Sample preparation – We’ll use AlbuVoid™ or other suitable enrichment product for sample preparation upfront to the 2-dimensional separation.
A protein sample of interest (the proteome) is first separated by wide range isoelectric focusing (IEF), followed by a modified second PAGE dimension to size separate proteins into resolved gel spots, without permanently compromising function. Depending upon the protein load and complexity, this will produce either 384 or 1536 resolved sub-proteomes. The separated proteins are then electro-eluted with the proprietary Protein Elution Plate (PEP), refolded, and transferred to a Master Plate for further enzyme assay. The enzyme activity/protein function is analyzed using a commercial kit or common reagents. A three dimensional profile of functional activity is produced from the starting proteome sample(s). These can be quantitatively compared and evaluated for different features, or can be followed by Mass Spectrometry for identification of the protein(s).
The PEP platform has demonstrated compatibility with:
- Proteases
- Protein Kinases
- Hexokinase Activity
- Oxidase/Reductase Family of Enzymes
- Alkaline Phosphatases
- Optional LC-MS/MS identification services can be provided.
A Final Report – A full report including methods, data analysis and client responsive goals is prepared.