Patent Application Describes New Proteomic Methods to Monitor Protease Inhibitor Function During Covid-19 Infections
Background
Serine Proteases
Serine proteases play critical roles in key biological processes including digestion, blood coagulation, and immunity. Most serine proteases, such as the prototypic enzymes trypsin and chymotrypsin, are secreted enzymes, while some such as Thrombin, are compartmentalized within intracellular granules, and released in response to stress or inflammation.
Other members of this family are cell membrane bound; the largest group being the Type II Transmembrane Serine Proteases (TTSPs). The defining features of TTSPs are an N-terminal transmembrane domain, tethered to a C-terminal
extracellular serine protease domain. TTSPs play an essential cooperative role during infectivity of influenza and coronaviruses, most especially in the cases of both the SARS outbreak of 2003 and the SARS-CoV-2 (Covid-19) pandemic. In both of these infections, the TTSP- TMPRSS2 cleaves the viral spike (S) protein, priming its binding to the receptor domain (ACE2) on the host cell, with subsequent fusion of viral and cellular membranes. With such a crucial role in influenza virus and coronavirus infections, antiviral therapeutic strategies targeting TMPRSS2 are now being investigated.
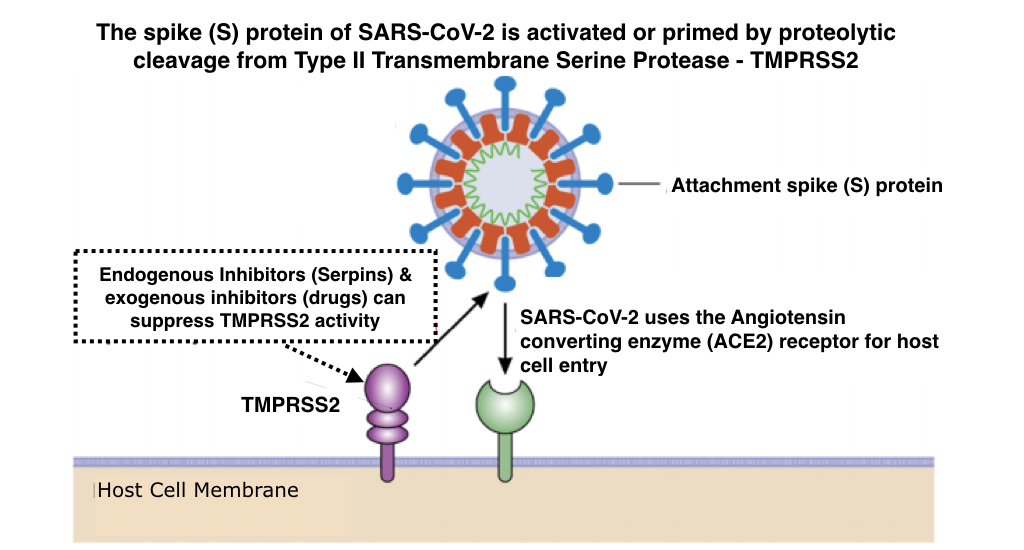
Although FDA-approved drugs that specifically inhibit TMPRSS2 are not yet available, some drugs that inhibit a variety of serine proteases have been approved for treatment of other diseases. These may also suppress influenza virus and coronavirus infections
1. So while repurposed drugs may offer treatment options, TTSPs can also be complexed by endogenous protease inhibitors in the general blood circulation and in biofluids. Especially noteworthy is that the Serpin family of protease inhibitors may be important regulators of TTSP inhibition2.
Serine Protease Inhibitors - also known as Serpins
Serpins can influence viral infectivity and severity in different ways. For example, plasminogen activator inhibitor 1 (PAI-1, SerpinE1) has been reported to efficiently inhibit TMPRSS2-mediated cleavage of influenza A virus surface glycoprotein HA and suppressed H1N1 influenza virus propagation ex & in vivo
3. Most importantly, lung- and plasma-derived α1-Antitrypsin (SerpinA1) has been shown to inhibit SARS-CoV-2 infection in cell lines and fully differentiated airway epithelium cultures, suggesting that this circulating inhibitory protein may suppress the spike priming process attributed to TMPRSS2, and serve a host protective role4-6. Furthermore, it will be important to investigate whether the levels of α1-Antitrypsin (SerpinA1) in the lungs or blood of COVID-19 patients inversely correlate with viral loads or disease progression4. In this context, a better understanding of the physiological relevance of α1-Antitrypsin (SerpinA1) and other Serpins in Covid-19 may be relevant in the development of new anti-Covid-19 therapeutic strategies and precision medicine biomarkers.
Challenge
Serpins have a very unique mechanism of action, one that is counter-intuitive to other inhibitory mechanisms. Because of this mechanism, there is no direct linear relationship between Serpin activities and conventional immunoassays to measure them. This can lead to egregiously misleading interpretations of Serpin function. Here’s why.
A "suicidal" family of protease inhibitors.
As proteolysis is irreversible, there is an essential balance and regulation of proteolytic cascades necessary to maintain normal homeostasis. In blood, this maintenance is regulated by the inhibitory Serpins in the general circulation, and collectively serve as central control for the innate immune response
7. With such importance, it is informative to highlight that Serpins exhibit conformational adaptabilities that result in opposing dualities of functional outcomes. Often called “suicidal”, inhibitory Serpins are in sharp contrast to the more conventional competitive mechanism for protease inhibitors that bind to and block access to the protease active site in a concentration dependent (Michaelis-Menten type) equilibrium. Such is not the case with Serpins, as the initial interaction starts through a decoy process (an intermediate Michaelis complex). Here, the protease sensing the Serpin as a suitable substrate, initially binds to a peptide region of the Serpin – known as the reactive center loop (RCL), that transiently protrudes from the core of the Serpin body. From this intermediate complex, one of two possible final outcomes are produced, called, respectively, the Substrate and Inhibitor Pathways, summarized as follows:
In the Inhibitor Pathway, the protease selectively cleaves a peptide bond in the reactive center loop (RCL) of the Serpin. During this process the active site is deformed, and hydrolysis cannot be completed. This process results in an irreversible covalent complex. Trapped in this suicidal covalent embrace,
both the protease and the Serpin are irreversibly modified and cannot be regenerated back to their active forms. In the Substrate Pathway however, the protease releases from the complex and remains active, but not so for the inhibitor as the RCL region is cleaved and the inhibitor becomes permanently inactive (the ‘Off’ subform)7-9.
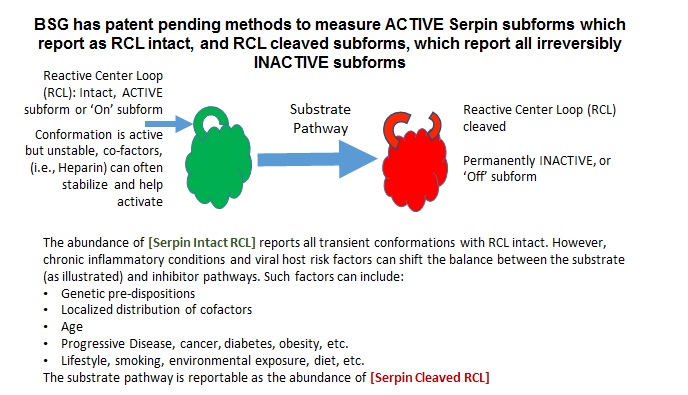
There are 10 major inhibitory Serpins reportable from blood and collectively account for about 5-10% of the protein mass in serum. With such a large regulating influence in blood, Serpins are themselves subject to many co-factors (e.g., Heparin) that act upon their inhibition efficiency. Any aberrations in this finely tuned mechanism can lead to the progressive loss of functionally active forms (the ‘On’ subform) or the accumulation of inactive forms (the ‘Off’ subform). Insufficient Serpin control of irreversible and inflammatory proteolytic activity can lead to systemic dysregulation of innate immunity and progression of disease, and can influence acute reaction to infectious insults. Conversely, activation of Serpin function therapeutically may partly explain the current clinical benefits seen for Heparin administration in some severe Covid-19 patients.
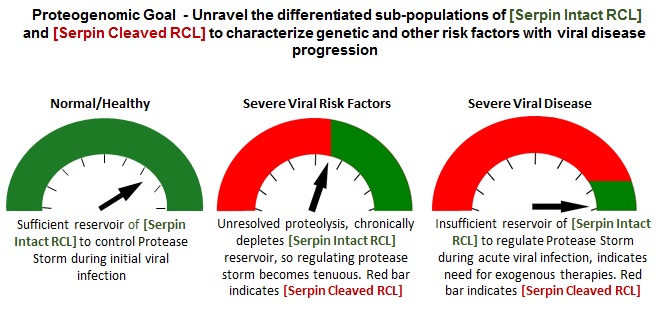
Notably, conventional immunoassays (i.e., ELISA) fail to report or represent in any way, the Serpin bifurcated functional pathways. As with all past methods, there is a mistaken assumption of a direct linear correlation between Serpin protein abundancy as measured by immunoassay, and associated functional potential. However, without better measurements for Serpin functionality, unresolved protease activity and consequential predisposition to severe disease upon exposure to viral infection, cannot be properly evaluated.
Solution
The patent disclosure describes new methods of proteomic analysis that can monitor how the body’s protease inhibitors are pre-disposed (or not), to suppress SARS-CoV-2 (Covid-19) viral entry, with potential for biomarkers to clinically manage severe disease and guide therapeutic development. The US provisional patent application is entitled “METHODS TO MONITOR FUNCTIONAL SUBFORMS OF SERINE PROTEASE INHIBITORS FROM BIOFLUIDS DURING VIRAL INFECTIONS”; Inventors: M. Kuruc & D. Roy, filed on October 27, 2020.
With viral infections, there comes an increased level of serine protease activity, due in part to both an exuberant innate immune response to counter the infectious insult, and the added TTSP activity that derives from the viral load and propagation. As a result, current clinical practice has no ability to predict and monitor the protease storm triggered by SARS-CoV-2 (Covid-19) and similar infections, and with that, any possible severity, thrombosis and other complications.
The purpose and scope of this patent application is to: >develop clinically actionable blood (and other accessible biofluids) biomarkers for viral infections that have a cell entry mechanism which utilize Type II Transmembrane Serine Proteases (TTSPs) as priming components, through proteomic measurement of functional subforms of the major inhibitory proteins that regulate protease storms - the Serpins, and >utilize one or more proteomic patterns from these Serpin subforms as biomarkers for precision medicine to clinically manage viral infections.
A proteomic solution is to report peptide features, derived from
ex vivo proteolysis (typically Trypsin) that can ascertain whether the RCL region of the Serpin is either intact or cleaved. This can be acquired by Liquid Chromatography coupled to Mass Spectrometry (LC-MS/MS), through either direct reporting features of the RCL region, or indirectly through peptide regions displaced upon cleavage of the RCL region. Sample prep products, in particular - AlbuVoid™ (depletes Albumin) and related AlbuVoid™ PLUS (depletes Albumin & IgG) also preferentially bind to intact proteins. This can be especially advantageous, serving to suppress the cleaved RCL peptide signal that would otherwise be observable, details provided upon request.
“This new provisional patent amplifies the proteomic methods we have developed to measure Serpin function by differentiating active vs. inactive Serpins from blood and other biofluids. While soluble serine proteases are released from systemic granulocytic cargo during the innate immune response, some viruses and especially SARS-CoV-2, utilize membrane bound serine proteases to gain entry into host cells and cause clinical pathology. In this context, a better understanding of the physiological relevance of endogenous inhibitors, like α1-Antitrypsin (SerpinA1) and other Serpins in Covid-19, will be exceedingly important as biomarkers to monitor the potential for severe disease, and in the development of new anti-Covid-19 therapeutic strategies. For this purpose, our new patent application will present a more complete picture of the functional profile of Serpin activity, and we welcome inquiries for its use.”, states Swapan Roy, Ph.D., President and Founder of Biotech Support Group.
Business Development:
For business development inquiries about our patent portfolio, please contact Matt Kuruc, 732-274-2866,
mkuruc@biotechsupportgroup.com.
References:
- influenza virus and coronavirus infections. Biochimie. 2017;142:1-10. doi:10.1016/j.biochi.2017.07.016.
- Hobson, John P., et al. "Mouse DESC1 is located within a cluster of seven DESC1-like genes and encodes a type II transmembrane serine protease that forms serpin inhibitory complexes." Journal of Biological Chemistry 279.45 (2004): 46981-46994.
- Dittmann, Meike et al. “A serpin shapes the extracellular environment to prevent influenza A virus maturation.” Cell vol. 160,4 (2015): 631-643. doi:10.1016/j.cell.2015.01.040.
- Wettstein, Lukas, et al. "Alpha-1 antitrypsin inhibits SARS-CoV-2 infection." bioRxiv (2020).
- Azouz, Nurit P., Andrea M. Klingler, and Marc E. Rothenberg. "Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV2-Priming Protease TMPRSS2." bioRxiv (2020).
- Oguntuyo, Kasopefoluwa Y., et al. "In plain sight: the role of alpha-1-antitrypsin in COVID-19 pathogenesis and therapeutics." bioRxiv (2020).
- Kuruc M, Zheng H, Sowerhardy A, Avadhani S, Roy D, et al. (2020) New Strategies to Categorize Blood for Proteomic Biomarker Discovery. Proteomics Bioinformatics, 2(2): 90-107.
- Law, Ruby HP (2006) An overview of the serpin superfamily. Genome Biology 7.
- Khan, Sazzad M (2011) Serpin inhibition mechanism: A delicate balance between native metastable state and polymerization. J Amino Acids 2011.
Other Resources
|