 |
 |
 |
 |
Hemoglobin Depletion Plus Protein Enrichment From Blood Card
- Hemoglobin voids in flow-through >98%, with <30 minute bind/wash/elute protocol
- Hemoglobin removal from whole blood lysates extracted from dried blood cards
- Hemoglobin removal from frozen and fresh whole blood
- Blood proteins and enzymes are enriched for potential biomarker and proteomic studies.
- Disposable, cost-effective hemoglobin depletion sample preparation protocol
- Removes hemoglobin from whole blood of diverse species including human, mice, sheep, bovine, goat, rat, etc.
Hemoglobin is a common contaminant from whole blood. The HemoVoid™ Blood Card protocol was designed to substantially reduce the presence of hemoglobin and its associated interference with many serum protein analytes. HemoVoid™ Blood Card is a silica based polyelectrolyte matrix, removes hemoglobin from dried whole blood card samples. The HemoVoid™ protocol uses mild buffers; the protocol conditions are so gentle that native enzyme activity is retained in elution fractions.
Click Here To View HemoVoid™ Blood Card
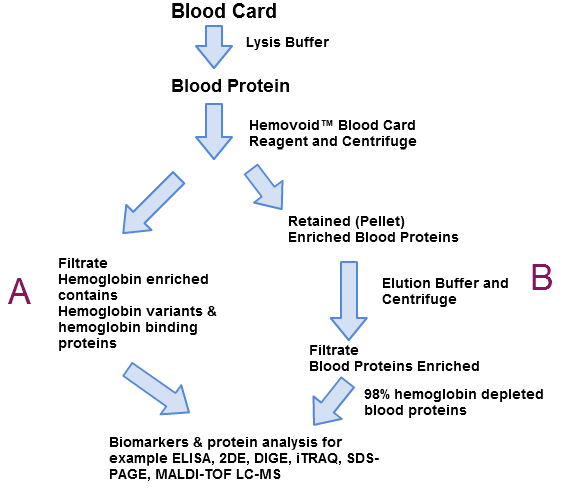
The HemoVoid™ Blood Card produces enriched proteins free of hemoglobin which are used for biomarkers and protein analysis(B). The hemoglobin enriched filtrate could have hemoglobin variants, hemoglobin binding proteins or other analytes optimal for biomarker studies(A).
|
Product
|
# of samples processed
|
Item No.
|
|
HemoVoid™ - Blood Card Reagent Kit
|
10 x 0.5" DBS preps
|
HVBC-10
|
|
HemoVoid™ - Blood Card Reagent Kit
|
50 x 0.5" DBS preps
|
HVBC-50
|
|
HemoVoid™ - Blood Card Reagent Kit
|
100 x 0.5" DBS preps
|
HVBC-100
|
Kit includes:
|
Kit Content
|
10 Prep
|
50 Prep
|
100 Prep
|
Reagent
|
|
HemoVoid™
|
0.5 gram
|
2.5 grams
|
5 grams
|
Supplied
|
|
Protein Extraction Buffer PEB
|
5 ml
|
25 ml
|
50 ml
|
Supplied
|
|
Binding Buffer HVBB, PH 6.0
|
15 ml
|
75 ml
|
150 ml
|
Supplied
|
|
Wash Buffer HVWB, PH 7.0
|
15 ml
|
75 ml
|
150 ml
|
Supplied
|
|
Elution Buffer HVEB, PH 9.8
|
3 ml
|
15 ml
|
30 ml
|
Supplied
|
|
SpinX Centrifuge tube filters
|
10
|
50
|
100
|
Supplied
|
Suggested Or Equivalent Supplier of Blood
Card: Whatman 903™ Protein Saver cards
|
|
|
|
Not Supplied
|
P. falciparum clone 3D7 cultured in human erythrocytes
Lasonder E, Green JL, Camarda G, Talabani H, Holder AA, Langsley G, Alano P.
The Plasmodium falciparum schizont phospho-proteome reveals extensive phosphatidylinositol and cAMP-Protein Kinase A signalling. J Proteome Research. 2012;
Authors Lasonder et al published an article titled,"The Plasmodium falciparum schizont phospho-proteome reveals extensive phosphatidylinositol and cAMP-Protein Kinase A signalling" in the journal of Proteome Research describing an overview of the Plasmodium falciparum phosphoproteome. Researchers discovered phosphorylated P. falciparum proteins and phosphorylation sites at the schizont stage of parasite development. Scientists found phosphorylation regulated DNA replication, transcription, translation and mitotic events (DNA packaging, chromosome organization, actin cytoskeleton). Researchers also found the CAMP-PKA signaling pathway to be involved in these events. The phosphorylation/dephosphorylation steps are important regulatory process of egress from and invasion into erythrocytes by merozoites. Scientists found phosphorylation of enzymes in the inositol pathway and discovered role of phosphorylation in merozoite egress and red blood cell invasion. Scientists developed an in vitro kinase assay and established the correlation of cAMP-PKA signaling for motility. CDPK1, GAP45, Myosin A were identified substrates with the glideosome motor complex. The analysis of Plasmodium falciparum schizont phospho-proteome involved steps which depleted hemoglobin from the samples using Hemovoid™ from Biotech Support Group. Freezing/thawing lysed P. falciparum schizont-infected erythrocytes and uninfected RBCs. Halt phosphase inhibitors (100X concentrated solution of sodium fluoride, sodium orthovanadate, sodium pyrophosphate and β-glycerophosphate) and protease inhibitor cocktail (AEBSF, aprotinin, bestatin, E-64, leupeptin, pepstatin A and EDTA) was added to the sample followed by centrifugation to separate the soluble and pellet fractions. Hemovoid™ successfully depleted hemoglobin from the soluble fraction yielding a hemoglobin depleted soluble fraction. Through the science discovered at Biotech Support Group, HemoVoid™ remains one of the only products in the market today that can separate hemoglobin from a blood sample, without destroying the intrinsic bonds within the protein. Mild elution maintains tertiary structure and simple transfer to secondary analysis.
Red Blood Cell Lysate
Barasa, Benjamin, and Monique Slijper. "Challenges for red blood cell biomarker discovery through proteomics." Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1844.5 (2014): 1003-1010
Biotech Support Group reports on a recent review article which describes the simplicity and efficiency of their proteomic sample preparation technology for selectively depleting hemoglobin, to help solve the dynamic range problem for comprehensive erythrocyte proteome analysis. The citation is: Barasa, Benjamin, and Monique Slijper. "
Challenges for red blood cell biomarker discovery through proteomics." Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1844.5 (2014): 1003-1010.In brief, this review describes the many challenges to generate in-depth RBC proteome analysis, such as to obtain pure red blood cells, and to acquire an in-depth proteome, despite the dynamic range problem due to a few highly over-represented RBC proteins – especially hemoglobin which accounts for approximately 97% of the cytosolic mass. The article states "Hemoglobin can also be depleted from an RBC lysate by employing Hemoglobind- [39] or HemoVoid [40] affinity systems. Hemoglobind consist of an elastomeric poly-electrolytic surface that has been optimized to bind Hb from serum samples with high affinity, and can as well be used to remove Hb from RBC lysates [39]. Walpurgis et al. used a complex matrix to deplete the RBC sample for Hb, named HemoVoid, which is made of a library of different ligand combinations, consisting of several kinds of ionic, aromatic, and polymer ligands [40]. Low abundance proteins in the RBC lysate are captured and enriched by the HemoVoid ligand library, while the high abundance proteins such as Hb and CA-I are thought to quickly saturate the system, and they primarily end up in the flow-through. The high abundance proteins in an RBC lysate can thus be easily separated from the low abundance protein fraction. The chosen Hb-depletion approaches by both Alvarez-Llamas et al. [39] and Walpurgis et al. [40] are well compatible with analysis of the RBC protein fractions by 1D or 2D gel electrophoresis, followed by protein identification through mass spectrometry.""It is worthwhile to note that the authors describe both our strategies for hemoglobin depletion, as the correct choice will vary with the application. With our own experience and with users such as those referenced in this article, we have gained the necessary knowledge to guide our users to the best option for hemoglobin depletion and/or low abundance enrichment" states Swapan Roy, Ph.D., President and Founder of Biotech Support Group.
References Acknowledged in the Review
1.[39] G. Alvarez-Llamas, F. de la Cuesta, M.G. Barderas, V.M. Darde, I. Zubiri, C. Caramelo, F. Vivanco A novel methodology for the analysis of membrane and cytosolic sub-proteomes of erythrocytes by 2-DE Electrophoresis, 30 (2009), pp. 4095–4108.
[40] K. Walpurgis, M. Kohler, A. Thomas, F. Wenzel, H. Geyer, W. Schanzer, M. Thevis
Validated hemoglobin-depletion approach for red blood cell lysate proteome analysis by means of 2D PAGE and Orbitrap MS Electrophoresis, 33 (2012), pp. 2537–2545
Hikosaka, Keisuke, et al. "
Deficiency of Nicotinamide Mononucleotide Adenylyltransferase 3 (Nmnat3) Causes Hemolytic Anemia by Altering the Glycolytic Flow in Mature Erythrocyte" Journal of Biological Chemistry(2014): jbc-M114.
Biotech Support Group reports on a recent research article which describes the simplicity and efficiency of their proteomic sample preparation technology for depleting hemoglobin, and enriching the low abundance proteome from human erythrocyte lysates. The article states “…hemoglobin was depleted…using a commercial kit (HemoVoid™). This crude protein-level pre-fractionation proved helpful in identifying additional proteins and N-termini”. In brief, the article describes a goal of the Chromosome-centric Human Proteome Project to identify all human protein species. With 3,844 proteins annotated as “missing” this is challenging. Enucleated and largely void of internal membranes and organelles, erythrocytes are simple yet proteomically challenging cells due to the high hemoglobin content (about 97% by mass) and wide dynamic range of protein concentrations that impedes protein identification. Using a N-terminomics procedure called TAILS, the authors identified 1369 human erythrocyte natural and neo-N-termini and 1234 proteins. From the HemoVoid™ treated, hemoglobin-depleted soluble fraction, 778 proteins were identified, 171of which were not represented in either the soluble non-depleted fraction or the membrane fraction. This study also establishes a general workflow suitable for the in-depth determination of the position and nature of human protein N termini in different tissues and disease states. The identification of 281 novel erythrocyte proteins and six missing proteins identified for the first time in the human proteome confirmed its utility. “While the authors acknowledged other low abundance enrichment methods, our HemoVoid™ product was chosen for protein level enrichment. I am pleased to see it proved exceedingly useful in this exciting new area of proteomic identification.” states Swapan Roy, Ph.D., President and Founder of Biotech Support Group.
Katja Walpurgis, Maxie Kohler, Andreas Thomas et al.
Validated hemoglobin-depletion approach for red blood cell lysate proteome analysis by means of 2D-PAGE and Orbitrap MS. Electrophoresis.2012;
This article states the HemoVoid™ process as a “…very efficient enrichment of low-abundant proteins by simultaneously reducing the hemoglobin concentration of the sample”, and that “…a two-dimensional reference map (pH 4-7) of the cytosolic red blood cell proteome was generated and a total of 189 different proteins were identified. Thus, the presented approach proved to be highly suitable to prepare reproducible high-resolution two dimensional protein maps of the RBC cytosol and provides a helpful tool for future studies investigating disease- or storage-induced changes of the cytosolic red blood cell proteome.” The identification of proteins in erythrocytes for proteomics studies is hampered by the huge abundance of hemoglobin (Hb) which hides other proteins and makes detection in 2-D gel-based separation difficult. It is therefore important to do pre-fractionation and depletion methods for the identification of low-abundant proteins. Authors Walpurgis et al demonstrate HemoVoid™’s application in the development of a protocol for proteomic analysis of hemoglobin-depleted RBC lysates in human blood from healthy donors. After using HemoVoid™, 2D-PAGE comparison of the unextracted hemolysate and the HemoVoid™ depleted hemolysate displays disappearance of the prominent and smeared hemoglobin ‘spot’ formerly containing high amount of hemoglobin on the gel is significantly reduced. Thus HemoVoid™ allows scientists to detect and study non-hemoglobin proteins using subsequent LC-MS/MS analysis for study of cytosolic proteome. Authors recorded that HemoVoid™ removed more than 98% of cellular hemoglobin, indirectly concentrated minor proteins and eliminated unbound hemoglobin. Upon comparison of the untreated and HemoVoid™ treated RBC lysates in the flowthrough and wash fractions by SDS-PAGE, scientists discovered the untreated RBC lysate showed two intense hemoglobin-derived bands (approximately 15 and 30 kDa), and the HemoVoid™-treated RBC lysate did show several bands which were not visible prior to hemoglobin depletion possibly representing non-hemoglobin proteins. Flow-through and three wash fractions contained some bands plus hemoglobin bands.
Mizukawa, B., George, A., Pushkaran, S. et al.
Cooperating G6PD mutations associated with severe neonatal hyperbilirubinemia and cholestasis. Pediatric Blood Cancer.2011;56: 840-842.
The paper titled, "Cooperating G6PD mutations associated with severe neonatal hyperbilirubinemia and cholestasis" uses HemoVoid™ for performing native gel electrophoresis and immunoblotting on blood samples from the patient and control subjects that were lysed and depleted of hemoglobin. Subsequent to using HemoVoid™, non-bound protein was eluted at pH 9.8 and placed in native sample buffer, pH 6.8 and analyzed by native gel electrophoresis in polyacrylamide gradient gels of 4–15%. Separating hemoglobin from erythrocytes by contacting erythrocytes with a hypotonic buffer solution at a rate sufficient to render the release of hemoglobin from said erythrocytes without significant lysis. The hemoglobin is then separated from the erythrocytes. Hemovoid™ allows for the purification of hemoglobin solutions of DNA, endotoxins and phospholipids by contacting the hemoglobin solutions with an anion exchange medium.
Sudha Neelam, David G Kakhniashvili, Stephan Wilkens et al.
Functional 20S proteasomes in mature human red blood cells Experimental Biology and Medicine.2011;236:580-591
Hemovoid™ is used to study the functional 20S and/or 26S proteasomes within red blood cells (RBCs; depleted of reticulocytes and leukocytes).Using methods such as double-immunofluorescence confocal microscopy to localize mature RBCs, proteasomes are isolated from mature RBC. After using Hemovoid™, a two-dimensional differential in-gel electrophoresis (2D-DIGE)approach was used to determine if proteasome-dependent protein degradation occurs within mature RBCs.
HemoVoid™: Hemoglobin Enrichment for Hemoglobin Variant Research
Biotech Support Group, LLC has been researching and creating innovative genomic and proteomic products for over 15 years. One of the new breakthroughs in our protein purification research is the novel method of separating and purifying hemoglobin from blood samples. Clinical researchers as well as lab technicians have been using Biotech Support Group's HemoVoid™ in various different applications in order to separate hemoglobin itself, from a milieu of proteins, in order to study the various antibodies, and biomarkers located on hemoglobin's surface. This is possible because hemoglobin retains its tertiary structure, and is not being denatured nor destroyed in the separation process. Through the science discovered at Biotech Support Group, HemoVoid™ remains one of the only products in the market today that can separate hemoglobin from a blood sample, without destroying the intrinsic bonds within the protein. Biotech Support Group's provides hemoglobin enrichment protocol from blood sample for hemoglobin variant research (HbS, HbF, HbA, HbA1c, Thalassemia, etc.)
Blood, Cytosolic Red Blood Cell Proteome
Walpurgis, Katja, et al. "Effects of gamma irradiation and 15 days of subsequent ex vivo storage on the cytosolic red blood cell proteome analyzed by 2D DIGE and Orbitrap MS." PROTEOMICS-Clinical Applications (2013).
Transfusion-associated graft-versus-host disease is prevented by gamma or X-ray irradiation of blood products. Radiation-induced dose-dependent erythrocytic damage causing hemolysis, storage/structural alterations, protein structure modifications, red blood cell (RBC) deformability and membrane leakage of extracellular K+ concentration. Sample preparation techniques involving hemoglobin depletion in addition to optimal irradiation dose and post-irradiation storage period is important for preventing transfusion-associated graft-versus-host disease and preserving blood product quality.
Cytosolic RBC proteome is researched using 2D-DIGE and nano-LC high-resolution/ high-accuracy Orbitrap MS. Sample preparation using HemoVoid™* for hemoglobin depletion ensures proper red cell concentrate preparation, storage and transfusion. Authors Walpurgis et al published an article in the journal Proteomics Clinical Applications titled, Effects of gamma irradiation and 15 days of subsequent ex vivo storage on the cytosolic red blood cell proteome analyzed by 2D DIGE and Orbitrap MS. Authors describe proteomic workflows and cite Biotech Support Group's HemoVoid™ for sample preparation. Whole blood samples containing citrate-phosphate-dextrose anticoagulant was collected. A leukocyte filter was used for leukoreduction. A Heraeus Cryofuge 6000i centrifugation was used for RBC, buffy coat and plasma separation.
A MacoPress blood component extractor. A colorimetric assay determined protein concentration of purified samples. Cytosolic RBC proteomic sample preparation involved cell lysis, hemoglobin depletion and protein determination. Washed RBCs were lysed, centrifuged and HemoVoid™ hemoglobin depletion protocol was performed. Upon removing hemoglobin, cytosolic proteins bound to HemoVoid™ matrix could be enriched. The elution fraction is centrifuged and concentrated. The retentate is washed with lysis buffer. 2D-DIGE was performed after gamma irradiation and ex vivo storage of the cytosolic RBC proteome.
Protein spots were identified via De-Cyder DIA module and ISTD images via BVA module. Altered protein spots and protein abundances were identified. The spotmaps of the untreated and unstored samples were compared with the samples stored following irradiation. In gel tryptic digestion and high-resolution/high-accuracy Orbitrap MS identified protein composition.Changes in protein abundances of the different samples was measured. Cytosolic RBC proteins, including TGase 2 or acylaminoacid-releasing enzyme, and the total number of identified peptides, sequence coverage, protein score was noted. 1D and 2D Western blotting in order to confirm their identity and validate the observed irradiation and storage-induced changes was performed. The 1D and 2D Western blots were validated against an antibody and the DDB1, VCP, and TGase 2 proteins were identified as sensitive markers for storage and irradiation-induced RBC lesions. Ionizing radiation or increased ex vivo storage which caused erythrocyte damage could affect RBC membrane. Potassium leakage, deformability and hemolysis are examples of RBC membrane defects. Moreover, gamma or x-ray radiation, irradiation dose, whole or RBC concentrate, pre-irradiation and storage before or after radiation was measured. Observed declines in protein abundance was measured. Irradiation-induced generation ROS was measured in RBC lysates as oxidation increased the intracellular proteases and insoluble protein aggregates also increased. Lactate dehydrogenase and hemoglobin were increased enhanced leakage from irradiated RBCs.
A decrease in cytosolic concentrations could be attributed to altered proteins codepleted from the cells. A screening assay which monitored RBC quality during ex vivo storage for lesions is developed using identified proteins as validated biomarkers. Engraftment and expansion of residual donor leukocytes could lead to transfusion-associated graft-versus-host disease. In blood transfusion recipients, irradiation of red blood cell (RBC) and ex vivo storage could benefit from hemoglobin depletion as proper sample preparation reduces variability in proteomic results. The development of a validated biomarker screening assay for quality of screening assays is an important field of research to prevent, detect and treat transfusion-associated graft-versus-host disease in blood transfusion patients.
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
References
Selection of HemoVoid™ References:
Red Blood Cells (RBCs) / Parkinson’s Disease / α-Synuclein
Cao, Chan, et al. " Deep learning-assisted single-molecule detection of protein post-translational modifications with a biological nanopore ." bioRxiv (2023): 2023-09. The authors demonstrate the ability of a biological nanopore, to detect and distinguish α-synuclein-derived peptides bearing single or multiple PTMs, occurring at different positions and in various combinations. To deplete Hemoglobin, the article states “Briefly, RBCs … treated using the HemoVoid kit, … to remove hemoglobin but also to enrich low abundant proteins such as α-synuclein.”
Klatt, Stephan, et al. " Optimizing red blood cell protein extraction for biomarker quantitation with mass spectrometry ." Analytical and Bioanalytical Chemistry (2020): 1-14.
The article describes the advantage of HemoVoid™ in detection of low abundance proteins when comparing their amounts (in percent) between four alternative extraction conditions, stating “… Most peptides, following HemoVoid™ extraction, showed ion abundances ranging between 1.00E+5 and 1.00E+6 (31%). In comparison to this, fewer peptides (10–23%) were within this range following extraction with all other protocols”. With respect to potential biomarkers for Parkinson’s Disease, the article states “For example, PRDX6 accounts for 0.4% of the total ion abundance after DOC (deoxycholate) extraction, whereas following HV ( HemoVoid™) extraction, this increases to 8%, a 20-fold enrichment”. The authors conclude that the HemoVoid™ method significantly reduces the concentration of hemoglobin, resulting in an increased signal-to noise of the remaining red cell proteins.
Elhadi, Suaad Abd, et al. "α‐Synuclein in blood cells differentiates Parkinson’s disease from healthy controls ." Annals of Clinical and Translational Neurology. The goal of this study was to determine whether blood cells expressing α-Synuclein can differentiate Parkinson’s disease (PD) from healthy controls. Two proteoforms - PSer129 a-Syn ( phosphorylated pathological form in Lewy bodies) and Oxidized a-Syn levels are observed in blood cells, but both at considerably lower concentration than total a-Syn, so the extremely high abundance of hemoglobin interferes with their analysis. To compensate, the article states for PSer129 α -Syn & Oxidized α -Syn detection by immunoassay, “ followed from hemoglobin clearance with HemoVoid kit”.
Vicente Miranda, Hugo, et al. " Posttranslational modifications of blood-derived alpha-synuclein as biochemical markers for Parkinson’s disease ." Scientific reports 7.1 (2017): 13713.
Posttranslational modifications (PTMs) in aSyn have been identified and implicated on its pathobiology. Since aSyn is abundant in blood erythrocytes, the study aimed to evaluate whether PTMs of aSyn in the blood might hold value as a biomarker for PD. As haemoglobin is the major protein component of erythrocytes lysates (90%), the article states “we depleted this … using HemoVoid…, enabling the additional concentration of other proteins of lower abundance. We confirmed the aSyn enrichment by immunoblotting (SDS-PAGE and dot-blot).”
Red Blood Cells (RBCs) / Other Applications
Mitra, Nibedita, et al. " Multi-Omics Analysis of Red Blood Cells Reveals Molecular Pathways Underlying Thalassemia Severity Beyond Globin Gene Mutations ." medRxiv (2025): 2025-02.
The study aims were to identify dysregulated molecular pathways in red blood cells contributing to thalassemia severity. In the methods section for Sample Preparation for RBC Proteomics Study, the article states “hemoglobin was depleted using the HemoVoid kit…”. This investigation finds six pathways which are responsible for thalassemia severity independent of mutational burden.
Wu, Na, et al. " Proteomic characteristics of plasma and blood cells in natural aging rhesus monkeys ." Proteomics: 2200049. This study sought to understand the aging process. For this purpose, the investigation analyzed and compared the protein expression spectrums in the blood of old and young rhesus monkeys. To extract blood cell proteins and deplete Hemoglobin, the article states “Blood cell proteins were lysed with…protein extraction solution (Bestbio, China)…After centrifugation…the supernatants were further depleted of Hemoglobin using HemoVoid™ .” Upon depletion, the study found 1183 proteins expressed differentially in blood cells.
Das, Sonu, et al. " A journey to unravel the pathophysiology of stable and exacerbated Chronic obstructive pulmonary disease through erythrocyte proteomics: A combined mass spectrometry/bioinformatics approach ." (2022). A label free relative quantification of erythrocyte cytosol proteome based on LC-MS/MS was performed on hemoglobin- depleted erythrocyte lysate samples of stable and exacerbated COPD, relative to healthy controls. To deplete Hemoglobin, the article states “ HemoVoid™, … from erythrocyte lysate samples to unmask low abundance…proteins… .”
Pawliński, Łukasz, et al. " Proteomic biomarkers in Gaucher disease ." Journal of clinical pathology 74.1 (2021): 25-29. For proteomics, quantitative analysis was performed by the isobaric tag for a relative and absolute quantitation (iTRAQ) method. The article states “Cells were lysed in lysis buffer (7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 1% DTT …, vortexed, incubated at 25 °C for 30 min and then centrifuged at 12 000 ×g for 15 min…the samples were purified using HemoVoid resin (Biotech Support Group) to remove haemoglobin contamination. The study found 31 proteins that significantly differed in concentration between GDt1 patients and a control group.
Christer, M. A. L. M., et al. " Methods for the detection of autologous blood-doping. " U.S. Patent Application No. 16/976,936. The patent application relates to the detection of autologous blood doping. More specifically, the invention relates to methods comprising tryptic digestion of samples of isolated red blood cell (RBC), specifically isolated RBC cytosol, followed by peptide mapping using LC-MS/MS. The invention’s description states “ Hemoglobin depletion was performed using HemoVoid resin … .”. Upon the preferable depletion of hemoglobin, the methods enable detection of increased levels of certain peptides in samples from subjects that have been subjected to autologous blood doping, compared to samples from non-doped control subjects.
Bollenbach, Alexander, et al. " GC-MS and LC-MS/MS pilot studies on the guanidine (NG)-dimethylation in native, asymmetrically and symmetrically NG-dimethylated arginine-vasopressin peptides and proteins in human red blood cells. " Journal of Chromatography B (2020): 122024.
Previous studies showed that human red blood cells are rich in large (> 50 kDa) asymmetric dimethylarginine -containing proteins of unknown identity. The study aimed to report the identity, biological activity and concentration of NG-methylated proteins by using GC-MS and LCMS/MS approaches. The article states “ we included in our method the use of HemoVoid™ to remove specifically most erythrocytic hemoglobin and to improve the SDS-PAGE separation of proteins for further processing. The HemoVoid™, … allowed removal of erythrocytic hemoglobin to a large extent from the hemolysate . … removal of hemoglobin by this technique enabled an effective separation by SDS-PAGE and isolation of bands… .”.
Kitao, Akihito, et al. " Band 3 ectopic expression in colorectal cancer induces an increase in erythrocyte membrane-bound IgG and may cause immune-related anemia ." International Journal of Hematology (2020): 1-10. Autoimmune hemolytic anemia (AIHA) is a rare comorbidity in colorectal cancer (CRC) and has an unknown etiology. To better understand cancer-related anemia, the authors’ investigated ectopic band 3 expression and erythrocyte membrane-bound IgG in a CRC cohort. To reduce the interference from Hemoglobin, the article states “Erythrocytes were lysed … and hemoglobin was depleted using HemoVoid ”.
Rosin-Arbesfeld, Rina, and Ronen SIMAN-TOV. "Article of manufacture and methods for increasing survival of red blood cells." U.S. Patent Application No. 15/739,857. The patent application describes an ex - vivo method of increasing survival of red blood cells (RBCs). The invention’s description states “The Haemolysates were enriched with over 95 % hemoglobin. For hemoglobin depletion , the hemoglobin depletion kit of HemoVoid … was used ”. Upon depletion of hemoglobin, a reduction in cytoplasmic actin levels was observed.
Nemkov, Travis, et al. " Hypoxia modulates the purine salvage pathway and decreases red blood cell and supernatant levels of hypoxanthine during refrigerated storage ." haematologica 103.2 (2018): 361-372. The goal of this study was to use proteomics in part to understand hypoxanthine catabolism in vivo for stored red blood cells. It is still unclear whether accumulation of hypoxanthine in stored red blood cell units is clinically relevant for transfused recipients. The article states “ Leukocyte-reduced human RBC from healthy donor volunteers were washed five times in phosphate-buffered saline prior to lysis in distilled water with sonication. Proteomic analyses of RBC membranes and cytosols were performed… RBC cytosolic proteins were depleted of hemoglobin using Hemovoid™ prior to high-pH reversed phase fractionation”.
Cortese-Krott, Miriam M., et al. "Identification of a soluble guanylate cyclase in RBCs: preserved activity in patients with coronary artery disease." Redox Biology (2017).
http://www.sciencedirect.com/science/article/pii/S2213231717306535
In brief, the authors aimed to investigate whether RBCs carry a functional soluble guanylate cyclase (sGC) signalling pathway and to address whether this pathway is compromised in coronary artery disease. The article states “ Using a commercial resin (HemoVoid™), which removes hemoglobin… and allows enrichment of soluble cytoplasmic proteins, we established a procedure that allows fast and reliable preparation of hemoglobin-free cell lysates from as little as 1-2 ml blood . In those samples, expression and activity of the cGMP-generating sGC, cGMP-hydrolyzing PDE5 and cGMP-transducing PKG was assessed by enzymatic assays and Western blot analysis”.
Feliciano, Amélia, et al. "Evening and morning alterations in Obstructive Sleep Apnea red blood cell proteome." Data in Brief (2017). http://dx.doi.org/10.1016/j.dib.2017.01.005
Using proteomics-based evaluation of red blood cells (RBC), the authors identified differentially abundant proteins associated with Obstructive Sleep Apnea Syndrome (OSA). Proteome variations between various time points were assessed. The article states “ RBC cytoplasmic fraction depleted of hemoglobin, using HemoVoid™ system, were analyzed by two-dimensional fluorescence difference gel electrophoresis (2D-DIGE), the 2D image software-based analyzed and relevant differentially abundant proteins identified by mass spectrometry (MS)”.
Philipp F Lange, Pitter F Huesgen, Karen Nguyen, and Christopher M Overall. ” Annotating N termini for the Human Proteome Project: N termini and Nα-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome”, J. Proteome Research., Just Accepted Manuscript • DOI: 10.1021/pr401191w • 21 Feb 2014. The article describes a goal of the Chromosome-centric Human Proteome Project to identify all human protein speciesUsing a N-terminomics procedure called TAILS, the authors identified from the HemoVoid™ treated, soluble fraction, 778 proteins were identified, 171 of which were not represented in either the soluble non-depleted fraction or the membrane fraction.
Katja Walpurgis, Maxie Kohler, Andreas Thomas et al. Validated hemoglobin-depletion approach for red blood cell lysate proteome analysis by means of 2D-PAGE and Orbitrap MS .Electrophoresis.2012;
Mizukawa, B., George, A., Pushkaran, S. et al. Cooperating G6PD mutations associated with severe neonatal hyperbilirubinemia and cholestasis . Pediatric Blood Cancer.2011;56: 840-842.
Sudha Neelam, David G Kakhniashvili, Stephan Wilkens et al. Functional 20S proteasomes in mature human red blood cells Experimental Biology and Medicine.2011;236:580-591
Red Blood Cells, Plasmodium extracts / Malaria
Machado, Patrícia Isabel Pires. Pyruvate kinase and glucose-6-phosphate dehydrogenase deficiencies and their association with malaria–population genetics and proteomic studies . Diss. Universidade do Porto, 2013.
Walpurgis, Katja, et al. " Effects of gamma irradiation and 15 days of subsequent ex vivo storage on the cytosolic red blood cell proteome analyzed by 2D DIGE and Orbitrap MS ." PROTEOMICS-Clinical Applications (2013).
Lasonder E, Green JL, Camarda G, Talabani H, Holder AA, Langsley G, Alano P. The Plasmodium falciparum schizont phospho-proteome reveals extensive phosphatidylinositol and cAMP-Protein Kinase A signalling . J Proteome Research. 2012;
Species Agnostic Applications
Lan, Qin, and Zhao-bing Gu. " Data-independent acquisition-based proteome profiling of red blood cells from dairy buffaloes under different types of heat stress ." Veterinary and Animal Science (2025): 100437. Heat stress (HS) induces hypoxia and oxidative stress, reducing animal health and livestock production. Red Blood Cell (RBC) lysates were isolated for data-independent acquisition-based proteomics to identify differentially expressed proteins involved in the HS response. The article states “ HemoVoid™ LC-MS On-Bead kit (Biotech Support Group, HVB-MS10) was used to remove Hb”. Digested peptides were analyzed by nanometric high performance liquid chromatography performed with a Q-Exactive HFX (Thermo Scientific) mass spectrometer, operated in DIA mode. Results showed that blood clotting factors, complement proteins, immunoglobulins, and vasoconstriction proteins were consistently decreased under the three types of HS conditions.
Puente-Marin, Sara, et al. "In Silico Functional Networks Identified in Fish Nucleated Red Blood Cells by Means of Transcriptomic and Proteomic Profiling ." Genes 9.4 (2018): 202. Label-free shotgun proteomic analyses were carried out for in silico functional pathway profiling of rainbow trout RBCs. The article states “ The cytosolic fraction, approximately 300 μL, was depleted of hemoglobin using HemoVoidTM kit (Biotech Support Group, Monmouth Junction, NJ, USA), in accordance with the manufacturer’s instructions” .
Nombela I, Puente-Marin S, Chico V et al. Identification of diverse defense mechanisms in trout red blood cells in response to VHSV halted viral replication F1000Research 2017, 6:1958 (doi: 10.12688/f1000research.12985.1)
The article states “ … LC ESI-MS/MS analysis of each of the fractions… . Briefly, the haemoglobin of the cytosolic fraction was removed using a column of HemoVoid™ kit…, following the manufacturer instructions ”.
For a full list of Hemoglobin Removal references, visit:
https://www.biotechsupportgroup.com/References-s/138.htm#hemoglobin-depletion
|
|
 |
 |
 |
 |


|
