|

The Potential for New Blood Biomarkers in the Management of COVID-19 Disease
Unleashing the Power of Proteomics To Better Understand the Innate Immune Response to Infectious and Non-infectious Inflammatory Stimuli
Background
Here is what the technical literature is saying about the immune response with COVID-19 patients.
“Significant evidence indicates that a dysregulated innate immune response contributes to the clinical presentation of patients with severe COVID-19 infections." J Exp Med (2020) 217 (6): e20200678.
https://doi.org/10.1084/jem.20200678
"Although limited by its observational nature, ...and indication bias, our findings suggest that systemic anticoagulation may be associated with improved outcomes among patients hospitalized with COVID-19." Journal of the American College of Cardiology. https://doi.org/10.1016/j.jacc.2020.05.001
“We identified molecular changes in the sera of COVID-19 patients compared to other groups implicating dysregulation of…, platelet degranulation and complement system pathways…” Cell 2020.
https://doi.org/10.1016/j.cell.2020.05.032
“Thrombotic complications seem to emerge as an important issue in patients with COVID-19. Preliminary reports on COVID-19 pandemic outcomes have shown that infected patients commonly develop thrombocytopenia (36.2%) and may have elevated D-dimer (46.4%), while these rates are even higher in patients with severe COVID-19 disease (57.7% and 59.6%, respectively).” Journal of Clinical Virology 127 (2020) 104362.
“In many respiratory diseases characterized by an intense inflammatory response, the balance between proteolytic enzymes (proteases, including elastases) and their inhibitors (proteinase inhibitors) is not neutral. Excess activity of neutrophil elastase (NE) and similar proteases has been reported to cause tissue damage and to alter the remodeling process in many clinical conditions such as pneumonia, respiratory distress, and acute lung injury.” CHEST, Volume 152, Issue 2, August 2017.
“The neutrophil-to-lymphocyte ratio (NLR) was identified as the independent risk factor for severe illness in patients with COVID-19 infection.” medRxiv 2020.02.10.20021584
“...studies suggest that at least a subset of severe COVID-19 infection involves a catastrophic, complement-mediated thrombotic microvascular injury syndrome with sustained activation… of complement.” Translational Research 2020.
All of this points to associations between severe COVID-19 disease and an overly exuberant innate immune response. When functioning properly, the purpose of the innate immune response is to immediately prevent the spread of pathogens throughout the body, and to initiate the second line of defense, the adaptive immune response (e.g., T Cells). So a normal resolution of the innate response leads to a productive handoff to the adaptive response. Conversely, an unresolved innate response may delay or confound a suitable adaptive response, as appears to be case with severe COVID-19 disease.
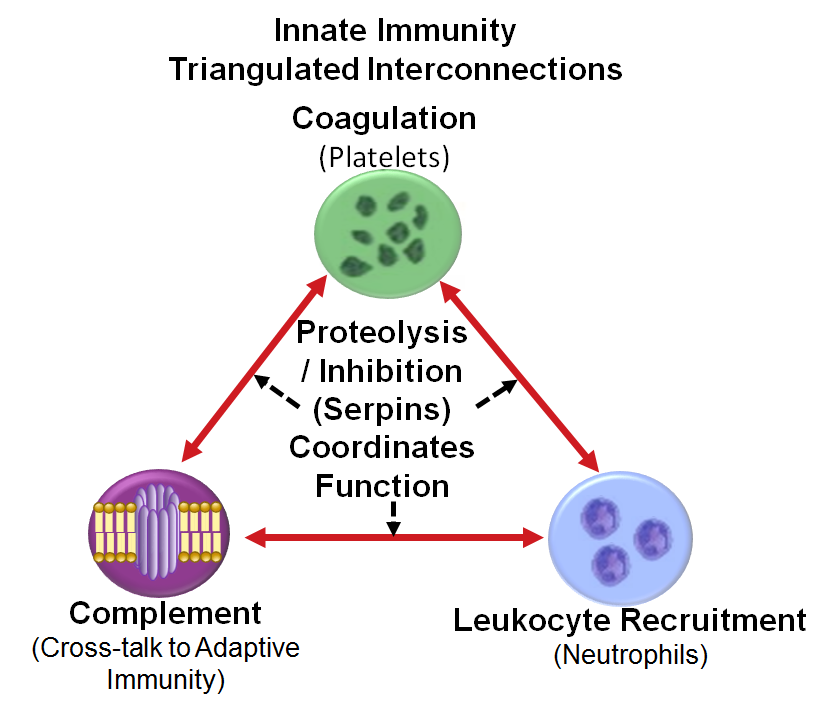
Because innate immunity needs to engage quickly, protein level orchestration is its foundation- see image. As observed now with anticoagulation therapy in COVID-19 patients, modulating even one rogue component can help to unwind dysregulation, and provide management of disease. For these reasons, we have focused on key regulatory elements that can be surveyed by proteomic analysis of serum/plasma including: Activated Neutrophils, Activated Platelets, & Activated Complement. Proteolysis and its regulation provides the central control function.
A disease state results whenever this triangulated proteome network becomes dysregulated via a confluence of infectious pathogens, heredity, lifestyle, or environmental stimuli.
- Blood mediates coordination between nonadjacent tissues, so it is essential to understand how this dysregulation manifests itself, regardless of the underlying causative factors.
- Quantitative proteomics from blood can help unravel these regulatory elements.
Challenge
The challenge of precision medicine is to develop and choose best available therapies based on actionable, accessible, and validated biomarkers. Current clinical practice has limited ability to predict and monitor COVID-19 patients with severe disease and presumed thrombotic complications. The biomarkers available, namely CRP, D-dimer, CBC, prothrombin time, and platelet count are very non-specific, and often report dysregulation only after severe disease occurs, and not before. So prognostic and precision medicine information from new biomarkers would be exceedingly valuable. At each stage of disease, biomarker profiling can:
- inform physicians as to the timing of intervention, and the preferred type, dose, and duration of treatment
- help companies develop future treatments, by finding clinically actionable blood-based biomarkers that can segment a disease population, to improve response.
Yet, extracting and characterizing functional changes and adaptations to disease for many of even the highest abundance proteins in circulation remains limited.
Solution
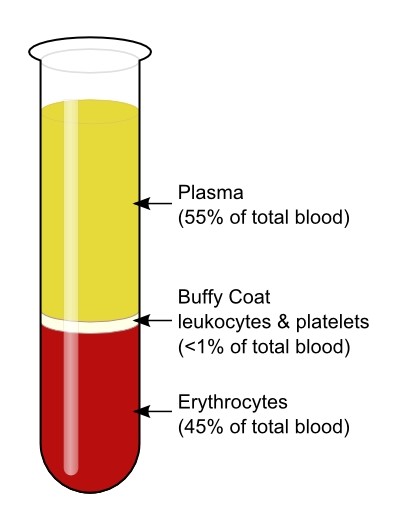 Proteomic information can be derived from all blood compartments
Proteomic information can be derived from all blood compartments
>Plasma/serum proteins and other circulating factors
directly regulate complex processes such as aging, the
development of chronic diseases, and severe acute disease
(i.e., acute respiratory distress syndrome).
>Activated leukocytes and platelets release granulocytic
cargo proteins in response to local inflammatory stimuli,
generating a protease storm, that if not resolved alters
steady state homeostasis contributing to both acute and
chronic disease.
>Erythrocytes do not have a nucleus, and being in continuous contact with complement proteins in the plasma, have a different make-up of complement regulators than nucleated cells. Increased complement activation or decreased complement regulation may result in dysfunction. This may explain some of the clinical symptoms- low oxygen levels and neurological manifestations, seen in severe COVID-19 patients.
BSG’s products and methods can help proteomic investigators explore all these blood compartments
Proteins are the ground truth of biology, but proteomic productivity can be improved
Proteomic productivity is not measured by Venn diagrams, but rather by quantitative
differences between proteins in samples representing a challenge or disease state, vs.
samples representing a normal or control state. However, small biological variances are hard
to measure robustly, given the nature of technical variance in LC-MS/MS analysis. Large
variance proteins (>2x) therefore are the best choices, but this limits the amount of potential
biomarker proteins to small numbers, maybe 10-20 rather than the anticipated 1000’s
expected when the concept of proteomics was first introduced. Aligned with that need, is also
the need to measure protein concentrations across several logs in one analysis.
In blood for example, the central Complement protein, C3 circulates at ≈1500 µg/ml, while
Complement Factor D circulates at ≈3 µg/ml. To measure both in one analysis requires that
the signal intensities at both ends of the spectrum be at least reasonably proportional to the
real concentrations, which at times can be across > 4 log ion abundance signal. Thus when
selecting potential biomarker proteins, it is best to bias towards proteins in the mid-high
abundance range, as low abundance proteins are often subject to signal to noise variance,
making them barely detectable and often well beyond the range where signal intensity is
proportional to concentration.
The solution demands:
- Fast and efficient sample prep products and methods to allow quantitative
monitoring of multiple blood proteins in the mid-high abundance range.
- Enrichment of biomarker proteins from low-abundance to mid-abundance to
improve linearity between the measurable peptide ion signals and true protein
abundances.
- Strategies for selection of biomarker proteins in the mid-high abundance range
which change characteristically with the onset of disease.
BSG has strived to address and service all these demands, and for that purpose we have
published two new resources.
Learn How
New categorization strategies for selection of innate immunity biomarker proteins in the
mid-high abundance range can change characteristically with the onset of disease.
Past proteomic analysis based on antigen recognition can lead to egregiously misleading
information on the status of protease regulation in the general blood circulation.
Unresolved inflammatory stimuli can lead to chronic disease and pre-dispose individuals
to a severe acute response to new infectious exposure.
The convergence of proteomic technologies has made the task immediately available to
understand the innate immunity proteome in the pathogenesis of pandemic infections,
cancer, and autoimmune disease.
Unbiased data-driven proteomic analysis can quickly lead to development of the next
generation of molecular tests for more precise and personalized treatment of patients.
For more information, click the links:
ebook Describes the Role of the Innate Immunity Proteome As a Central Modulator
of Disease from Pandemic Infections to Cancer
Journal Article Describes New Strategies to Categorize Blood for Proteomic
Biomarker Discovery
|