Albumin and High Abundance Depletion
Diverse Albumin Depletion and Enrichment Technologies Enhance Efficiency and Simplicity of Achieving Quality Proteomic Information
By combining two supportive enrichment strategies, BSG offers distinct and diverse technologies to help researchers selectively enrich or bind, in order to achieve quality results.
Background
Biotech Support Group develops cost effective and efficient sample prep methods essential for expanding proteomics into routine healthcare.
Enrichment/Depletion is one of the key BSG Advantages. Leading the way in proteomic sample prep BSG offers a choice of two strategic technologies for proteome enrichment: by either selectively binding high abundance or enriching- choosing not to bind- in order to get the necessary results.
The Challenge
Many proteomic enrichments often use immuno-affinity depletion to remove one or more high abundance proteins from plasma or serum. Some common limitations of this challenge include high costs, regeneration requirements which may result in a diminished and inconsistent performance, as well as a required marriage of species to antibody. Because of these limitations, researchers need ways to enrich differently.
The classical plasma proteins generally fall into functional categories, forming the vast majority of the mid-to-high abundance proteome. In serum, these sub-proteomes by mass content are:
- Albumin 50-60%
- Immunoglobulins 10-20%
- Transport Proteins (Transferrin, Apo) 5-10%
- Complement related Proteins 3-5%
- Protease Inhibitors 2-5%
- All others 2-5%
While these sub-proteomes are required for normal body homeostasis, they nevertheless can become dysfunctional during acute-phase and chronic stimuli. In some cases, enriching certain categorical proteomes can provide mechanistic insight into the disease pathology.
Depending on the needs of the investigation, it can be valuable to consider that one or more of these categorical sub-proteomes is simply background noise that needs to be depleted. While in other cases, these same categorical sub-proteomes might provide new data and information and consequently, should not be depleted. One such categorical sub-proteome is the complement system.
The complement system is a biochemical cascade made up of approximately 20 soluble plasma proteins along with membrane-bound receptors, and it represents a major part of the innate immune system. Orchestrated to rapidly defend the host against a variety of invading microorganisms, the complement system extensively communicates with associated biological pathways ranging from immunity and inflammation to coagulation, highlighting its importance in the activation and control of the generalized immune response.
Nevertheless, complement dysregulation has been implicated in many chronic diseases including Alzheimer’s, macular degeneration, atherosclerosis, and cancer. Characterizing complement on a proteome scale can therefore offer new discovery strategies for biomarkers of disease.
The role of BSG’s proteome separations can help in this effort.
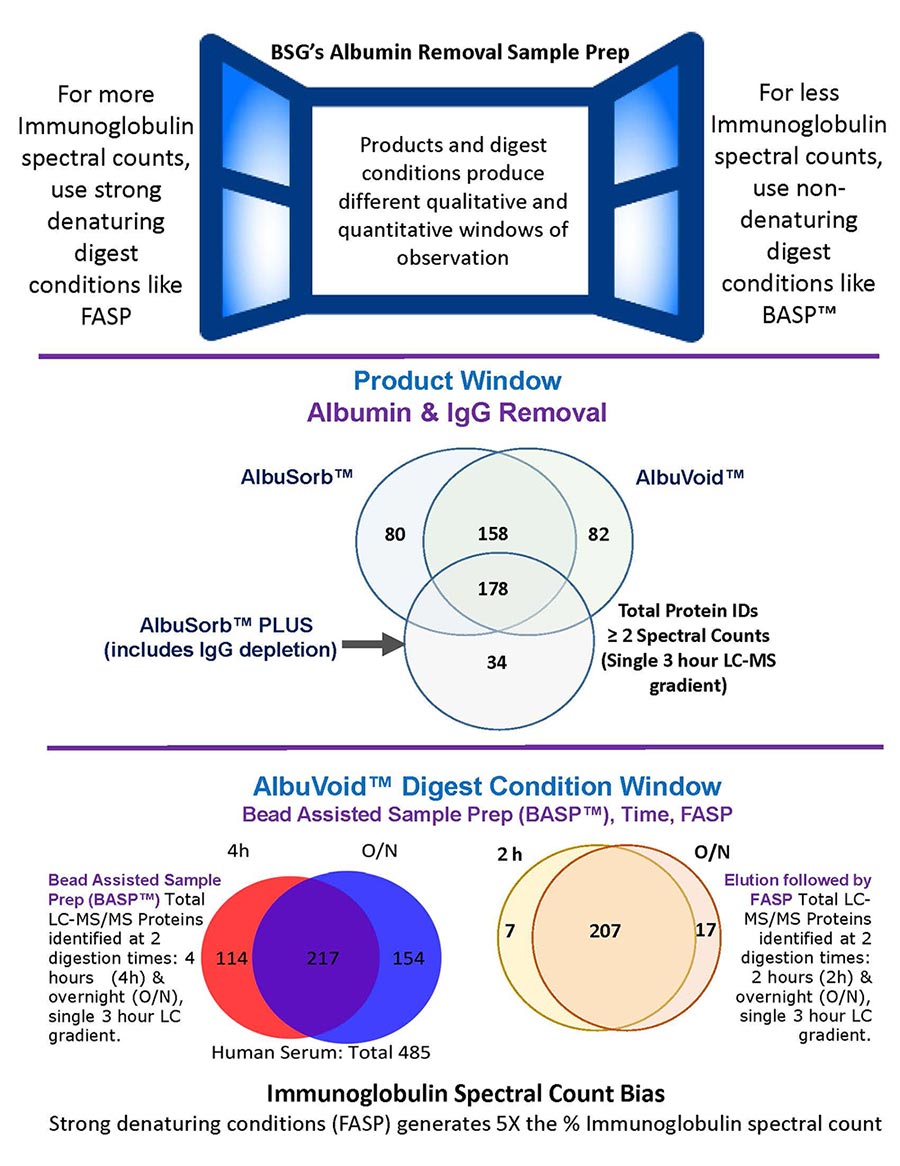
The Solutions: AlbuVoid™ and AlbuSorb™ PLUS
The error-prone disadvantages and challenges reviewed above are largely avoided by methods and developments supporting BSG’s AlbuVoid™ and AlbuSorb™ PLUS Albumin removal products. Without the use of immuno-affinity,
AlbuSorb™ PLUS selectively binds Albumin, Immunoglobulins and complement. And, AlbuVoid™ works inversely, binding the underlying sub-proteome to the beads, and voiding Albumin in the unbound fraction; Immunoglobulins and complement are recoverable.
The Outcomes
Products and digest conditions produce different proteome qualitative and quantitative windows of observation. This can be especially valuable to bias towards or against certain families of proteins. Here we report the serum proteome bias characteristics of AlbuVoid™ & AlbuSorb™ PLUS, using LC-MS (singular 3 hour gradient) reporting metrics.
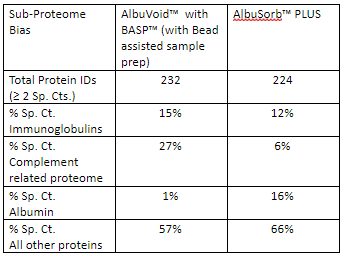
As can be seen from the Table above:
- AlbuVoid™ very efficiently enriches the complement and Immunoglobulin related sub-proteomes.
- AlbuSorb™ PLUS very efficiently depletes the complement and Immunoglobulin related sub-proteomes.
Proteomic characterization of complement can be done by a variety of analytical platforms including, protease activity, ELISA, Western blot and cellular response. Through the use of either of these two solutions, researchers will be equipped to better improve outcomes, regardless of the analytical platform.
Related References
BSG’s Albumin Removal Sample Prep and Bias Characteristics
Mihara, Keisuke, et al. "
Identification of Specific Protein Markers of Rheumatoid Arthritis in Synovial Fluid and Serum." Journal of Hard Tissue Biology1 (2018): 55-58.
David L. Wang, Chuanguang Xiao, Guofeng Fu, Xing Wang and Liang Li.
Identification of potential serum biomarkers for breast cancer using a functional proteomics technology. Biomarker Research (2017) 5:11. DOI 10.1186/s40364-017-0092-9
Zheng H, Zhao C, Qian M, Roy S, Arpa A, et al. (2015)
AlbuVoid™ Coupled to On-Bead Digestion – Tackling the Challenges of Serum Proteomics. J Proteomics Bioinform 8: 225-230.
Swapan Roy and Matthew Kuruc. "
The Functional Subproteomes of Serpin Protease Inhibitors are Now Open for LC-MS Biomarker Discovery." MOJ Proteomics Bioinform 3.6 (2016): 00106.
Swapan Roy, Amenah Soherwardy, Ravish Amin and Matthew Kuruc.
AlbuSorb™ Product Extension combines Albumin and Immunoglobulin Depletion in a Consumable Format. Poster reprint first presented at 12th Annual US HUPO 2016 Conference, held March 13 – 16, 2016 Boston, MA, USA
Happonen KE, Fürst CM, Saxne T, Heinegård D, Blom AM.
PRELP Protein Inhibits the Formation of the Complement Membrane Attack Complex. The Journal of Biological Chemistry. 2012;287(11):8092-8100. doi:10.1074/jbc.M111.291476.
|