Sample Prep for Mass Spectrometry
High abundance proteins and lipids are well known interferences in mass spectrometry based proteomic analysis. For years the protein depletion toolkit was limited primarily to immuno-affinity chromatography and other biologically-derived tools. BSG offers two complementary technologies for proteomic researchers to selectively bind or enrich, in order to achieve the best results. By using consumable non-biologically derived beads, researchers will be able to improve efficiency and quantitation essential for expanding mass spectrometry into routine healthcare. AlbuVoid™ LC-MS On-BeadAlbumin depletion plus low abundance protein enrichment, coupled with optimized on-bead digestion (BASP™) protocols for LC-MS serum and plasma proteomics
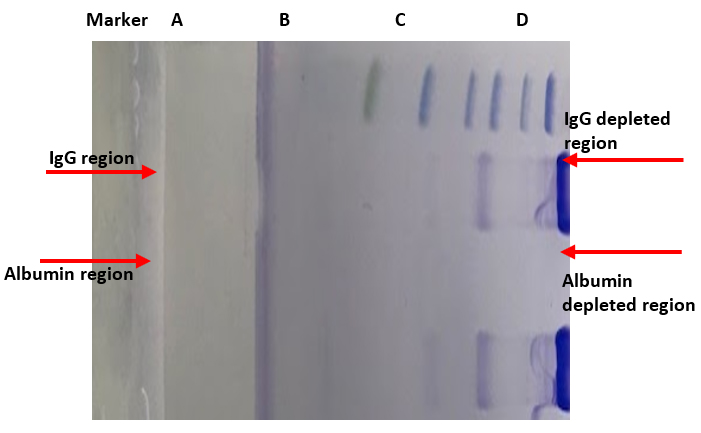
AlbuVoid™ PLUSAlbumin and IgG Depletion From Serum/Plasma for Proteomics
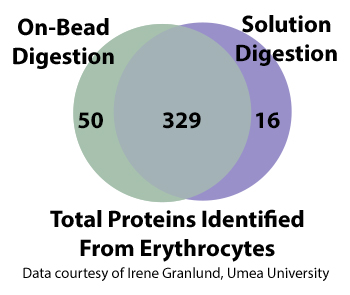
HemoVoid™ LC-MS On-BeadHemoglobin depletion plus low abundance protein enrichment, coupled with optimized on-bead digestion protocols (BASP™) for LC-MS erythrocyte & blood proteomics
AlbuSorb™Selectively Binds Albumin
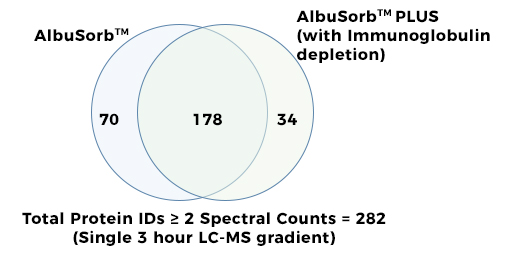
AlbuSorb™ PLUSSelectively Binds Albumin & Immunoglobulin
Quantitative Bias Comparison between
ReferenceFor serum exosome enrichment
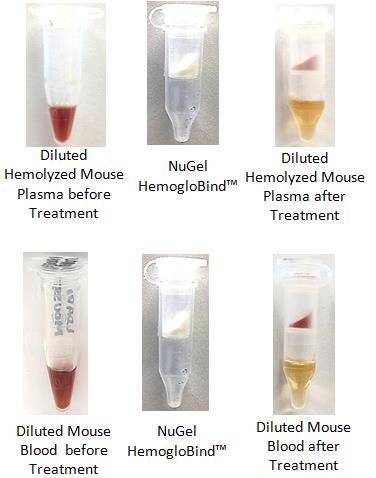
HemogloBind™
|

- About
- Products
- Hemoglobin Removal Kits
- Lipid Removal & Clarification
- Urine Protein & Low Abundance Enrichment
- Class Specific Enrichment
- Sample Prep Mass Spectrometry
- Functional & Chemical Proteomics
- Genomic Sample Prep
- Accessories
- Technical Resources
- References
- Publications & Reports
- FAQs
- Case Studies
- Cleanascite™ Unlocks Insights into Lipid-Driven Tumor Immunosuppression
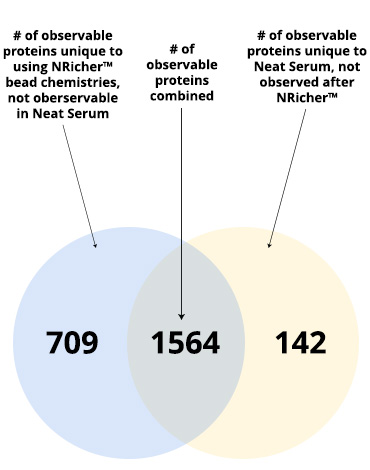
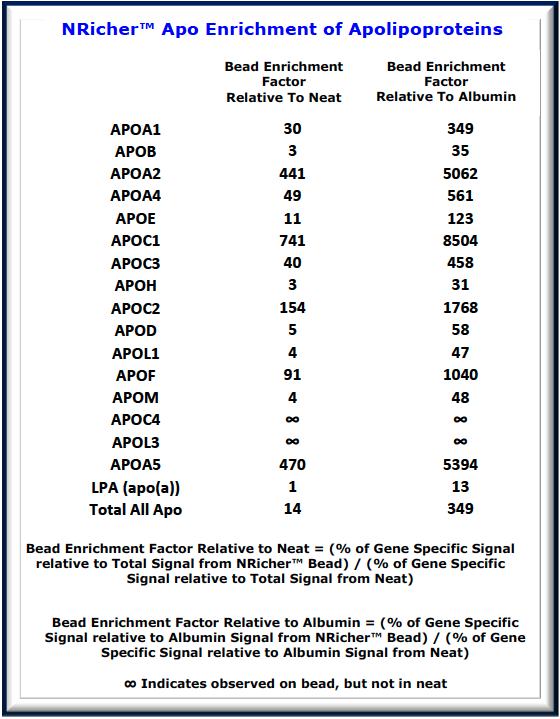
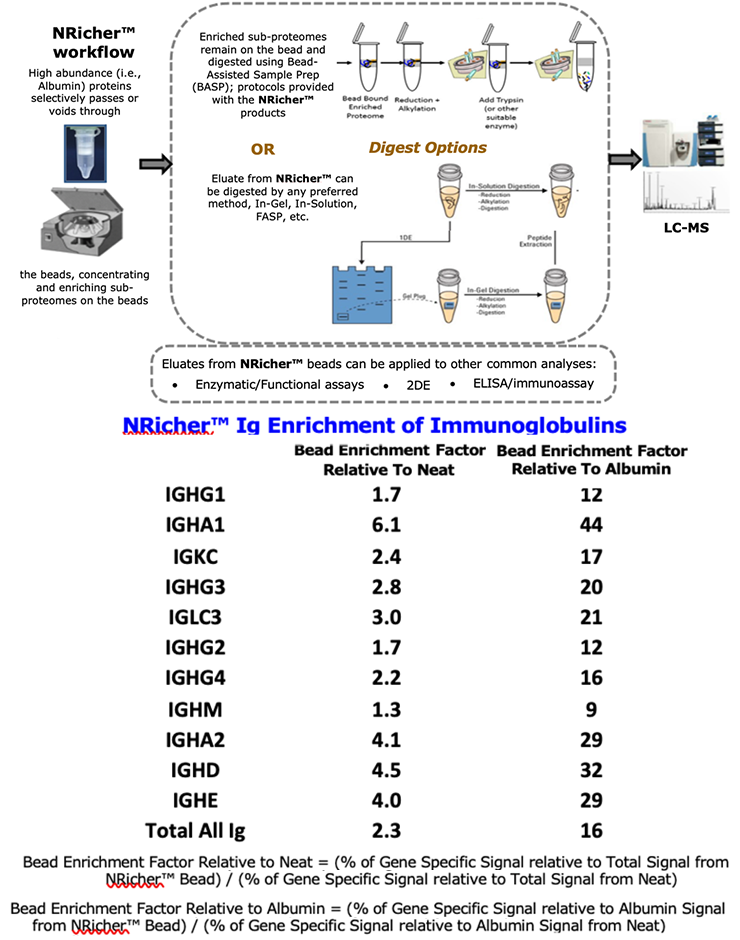
- NRicher™ Bead Platform Provides Unique Sub-Proteome Biases And Fit For Purpose Opportunities for Targeted LC-MS Quantification
- BSG Products To Assist in Analyzing Macrophage Polarization
- Ectodomain Shedding and Enrichment of the Soluble Membrane Proteome
- Investigate out of the Venn Diagram box
- Methods to selectively deplete or purify Hemoglobin from Dried Blood Spots (DBS)
- The Utility of HemoVoid™ is Demonstrated in 3 Proteomic Investigations Identifying Potential Disease Specific Biomarkers
- The 4 common features of our sample prep products, known as the BSG Advantage, are highlighted in a selection of journal references.
- AlbuVoid™ Workflows Advance Cell Secretome Proteomics
- Lipid Removal for Phenotypic Cell Response in Cancer Research
- The Influence of Sample Prep Bias on LC-MS Targeted Peptide Quantification in Serum Proteomics
- Re-imagining proteomics for developing precision medicine biomarkers of the innate immune response in SARS-CoV-2
- Patent Application Describes New Proteomic Methods to Monitor Protease Inhibitor Function During Covid-19 Infections
- Efficient Hemoglobin Removal Advances Red Cell Proteomics Offering Many New Insights Into Inflammation and Infectious Disease
- The Potential for New Blood Biomarkers in the Management of COVID-19 Disease
- Establishing the Utility of HemoVoid™ and HemogloBind™ as Enrichment Tools for Proteomic Analysis of Red Cells and Whole Blood in Parkinson’s Disease
- Species Diversity Supported By BSG Products
- Poster Report Describes Loss of Functional Serpin Activity In Cancer Patient Blood
- AlbuVoid™️ PLUS & AlbuSorb™️ PLUS Evaluating Different Windows of Observation Solves The Many Challenges of Serum Proteomics
- Tackling the Challenges of Serum Proteomics
- Lipid Removal Sample Prep for Cell Response Applications
- Sample Prep for Proteomic Analysis of Saliva
- Biotech Support Group Featured in Book, "Functional Proteomics – Methods and Protocols"
- Sample Prep Liquid Biopsy Products Suitable for Proteomic Profiling of a Variety of Body Fluid Sample Types
- Albumin and High Abundance Depletion
- Using HemogloBind™ as a Hemoglobin Binding Reagent
- Diverse technologies available for researchers to selectively bind or enrich exosomes and extracellular vesicles.
- Stroma Liquid Biopsy™ Biomarkers Profile Pan-Cancer Dysregulation of the Serum Proteome
- Diverse Depletion and Enrichment Technologies Enhance Simplicity and Efficiency of Obtaining Quality Proteomic Information
- Use On-Bead Digestion to Improve Time Required for Serum Digestion
- Using AlbuVoid™ as a Serum Protein Enrichment Kit in Functional Proteomics
- Using Cleanascite™ as a Lipid Absorption and Clarification Reagent
- Using HemoVoid to Remove Hemoglobin Before Analysis
- Blog
- Contact
- Liquid Biopsy