AlbuSorb™ Applications
Urine
Zubiri, Irene, et al. "Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis." Journal of proteomics 96 (2014): 92-102.
Urine exosome isolation via ultracentrifugation followed by albumin depletion provides researchers with knowledge of renal regulation and could lead to the identification of biomarkers for diabetic nephropathy and diabetic mellitus. By removing albumin using AlbuSorb™ authors identified more urinary proteins such as flotillin-2, lamp-1, PODXL, tsg-101 from exosome fractions of urine samples. Proteinuria in urine samples causes contamination of urine samples for proteomics research. Sample preparation protocols with depletion could complement precipitation techniques as less contamination is present in exosomal fractions upon depletion, enrichment, concentration and sample clarification. Urine exosome isolation via ultracentrifugation upon performing depletion provides researchers with knowledge of renal regulation and could lead to the identification of biomarkers for diabetic nephropathy and diabetic mellitus. Upon depletion of albumin, urine exosomes from diabetic and health controls were analyzed by LC-MS/MS and selected reaction monitoring (SRM). The research cites AMBP, MLL3, VDAC1 as proteins in urinary exosomes of diabetic nephropathy patients.
Cerebrospinal Fluid
Gwenael Pottiez, Pawel Ciborowski. Proteomic Profiling of Cerebrospinal Fluid Expression Profiling In Neuroscience. Neuromethods.2012;64:245-270
Authors Pottiez et al published a chapter in the book Expression Profiling in Neuroscience, Neuromethods titled, Proteomic Profiling of Cerebrospinal Fluid, on proteomic profiling platforms which analyze cerebrospinal fluid (CSF) for protein biomarkers and developing protein profiles of CSF for early identification of neurological diseases. Authors provide examples of affinity-based systems for removing most abundant proteins and cite AlbuSorb™ albumin depletion kit from Biotech Support Group. Moreover variations of protein concentration yielded by immunodepletion of CSF samples from nondemented (ND) patients and patients with HIV-associated dementia (HAD) are recorded. Detailed protocols are provided on analysis of:
Protein based profiling of intact proteins in CSF: Gel analysis by two-dimensional gel electrophoresis with DIGE technology and protein identification by tandem mass spectrometry of in-gel-digested protein spots. Next database searches, statistical analysis and validation follows.
Synovial fluid
Happonen, K. E., Fürst, C. M., Saxne, T., Heinegård, D., & Blom, A. M. (2012). PRELP protein inhibits the formation of the complement membrane attack complex. Journal of Biological Chemistry, 287(11), 8092-8100.
Approximately 65% of the total protein in normal synovial fluid consists of human serum albumin. Authors Kaisa E. Happonen et al published an article titled, "PRELP inhibits the formation of the complement membrane attack complex" in the Journal of Biological Chemistry which cites AlbuSorb™ from Biotech Support Group for albumin depletion from synovial fluid for detecting and measuring Proline arginine rich end leucine-rich repeat protein (PRELP). After using AlbuSorb™, synovial fluid proteins on two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) is studied. Examples of elevated biomarker candidates which are elevated in erosive or nonerosive rheumatoid arthritis (RA) are G3PDH,Peptidylprolyl isomerase, Cystatin B, Phosphoglycerate mutase 1, α2-plasmin inhibitor, S100A8 (calgranulin A), IgG1H Nie, Thymosin-β4. The ultimate goal of such application would be to comparatively find and analyze novel biomarkers of rheumatoid arthritis and search for the same biomarkers in synovial fluid of disease and non disease patient samples by proteomic techniques.
Serum
Swapan Roy, Matthew Kuruc. The Functional Subproteomes of Serpin Protease Inhibitors are Now Open for LC-MS Biomarker Discovery. MOJ Proteomics Bioinform 2016, 3(6): 00106
The authors consider that the conformational variants of the unique family of protease inhibitors annotated as SERPINs, are most often underrepresented in proteomic analyses. This limits understanding the complex regulation that this family of proteins presents to the networks within the protease web of interactions. Using bead-based separation provided by the NuGel™ family of proteomic enrichment products - notably AlbuVoid™ & AlbuSorb™, the authors demonstrate their utility to satisfy investigations of serum SERPINs. The authors also suggest their use to develop functional profiles of the SERPIN Proteoform, and how those can establish relationships to disease phenotypes, gene mutations, and deregulated mechanisms.
Holmberg, Rebecka, Essam Refai, Anders Höög, Rosanne M. Crooke, Mark Graham, Gunilla Olivecrona, Per-Olof Berggren, and Lisa Juntti-Berggren. "Lowering apolipoprotein CIII delays onset of type 1 diabetes." Proceedings of the National Academy of Sciences 108, no. 26 (2011): 10685-10689
The glycoprotein Apolipoprotein C-III (apoCIII) inhibits lipolysis and its expression is documented to play a vital role in the development of hypertriglyceridemia when increased. The aim of this study was to determine apoCIII’s increased levels in serum in T1D patients and that it affects the function and survival of pancreatic β cells in vitro. AlbuSorb™ from Biotech Support Group is used to remove albumin from rat serum samples. Scientists setup experiments implementing the animal model diabetes-prone BB rat (DPBB) to ascertain if apoCIII increases contributed to calcium increase and B-cell death in vivo. To evaluate the levels of apoCIII in pos, neg, and control sera, scientists used Albusorb™ (Biotech Support Group) to remove albumin from serum samples. Proteins from the freeze-dried collected samples were eluted and dissolved in 100 μL 0.1% TFA. Area under curve on an ACE C18 10- × 0.21-cm column 20–60% where apoCIII elutes evaluated and ApoCIII was identified by MALDI mass spectrometry. Scientists discovered that treating prediabetic animals with an antisense against apo CIII prolonged diabetes onset.
Tang MX, Ogawa K, Asamoto M.
Effects of Nobiletin on PhIP-Induced Prostate and Colon Carcinogenesis in F344 Rats Nutrition and Cancer.2011;63(2):227-33
Authors showed how 0.05% citrus flavonoid nobiletin inhibited PhIP-induced rat prostate and colon carcinogenesis. AlbuSorb™ was used for albumin depletion from serum samples to enable leptin expression. Following this, serum samples were diluted and denatured in the presence of sodium dodecyl sulfate and 2-mercaptoethanol by heating at 100◦C for 5 min. Then, proteins in each sample were electrophoretically. To prevent, nonspecific binding on the membranes 5% skim milk at room temperature for 1 h, followed by incubation with a polyclonal rabbit antileptin antibody allowed for leptin expression by western blot.
Holmberg, Rebecka
Apolipoprotein CIII and Ljungan virus in diabetes 2010. Doctoral Thesis
Lowering the levels of apolipoprotein CIII is beneficial to prevent development of type 1 diabetes. Albusorb™ was used on isolated islet cells from sera in prediabetic rats undergoing antisense treatment. Albusorb™ removed >90% albumin from serum samples. The method involved adding serum to a binding buffer with AlbuSorb™ powder followed by mixing and centrifugation. The supernatant was collected, freeze dried in 100ml 0.1% TFA and run on ACE C18 column 20-60%. The apolipoprotein CIII elutes were analyzed with by area under the curve measurements. Analysis of apolipoprotein CIII was done using MALDI mass spec.
Lu Q, Zheng X, McIntosh T
Development of different analysis platforms with LC-MS for pharmacokinetic studies of protein drugs. Analytical Chemistry.2009;81(21):8715-23
Using AlbuSorb™'s albumin depletion method first and then digest the depleted albumin solution (flow through fraction) for the subsequent LC-MS analysis of peptides, either 1-dimensional LC or 2-dimensional LC (ion exchange and reversed phase) with MS analysis. In this paper, authors use AlbuSorb™ from Biotech Support Group in a sample of serum (i.e., 30 μL) containing the protein drug along with a binding buffer provided (i.e., 250 μL) and then 40 mg of AlbuSorb™ powder is added in a spin-tube. At room temperature, the sample was mixed for 5-10 min on a rotating shaker, the spin-tube was centrifuged for 2 min, and the supernatant was collected for further analysis.
AlbuVoid™ Applications
Serum
MONMOUTH JUNCTION, NJ -- Biotech Support Group reports on a new application on AlbuVoid™, an albumin depletion reagent that selectively voids albumin and enriches the low abundance proteome. The application report obtains unique protein identification’s from human and rat sera upon depleting albumin using AlbuVoid™ in addition to providing a serum proteomic workflow. Subsequent to using AlbuVoid™ isobaric labeling, glycopeptide enrichment with Concanavalin-A (ConA) 4B, PNGase treatment to remove glycosylation, liquid chromatography mass spectrometry (LC-MS/MS) and computational peptide or protein annotation is possible. The AlbuVoid™ proteomics workflow on albumin depletion has compatibility with quantitative label (i.e., isobaric tags for relative and absolute quantitation (iTRAQ)) and label-free LC-MS methods. The application report provides data on immobilized ConA to enrich glycopeptides and enzymatic cleaving of glyco-bond producing glycoprotein fraction specific peptides. On bead digestions minimize proteolytic hydrolysis inconsistencies and more glycoproteins in total protein sample are observed. In-gel digests, solution digests and C18 desalting are not required and minimum mis-cleavages occur.
Download the application report and obtain the serum proteomic workflow using AlbuVoid™ on Biotech Support Group website.
Kuruc Matt "Application Report -
AlbuVoid™ & On-Bead Digestion: Tackling the challenges of serum proteomics."
Discovery of Functional Serum Biomarkers Using AlbuVoid™ Enrichment and the ArrayBridge PEP Profiling Platform.
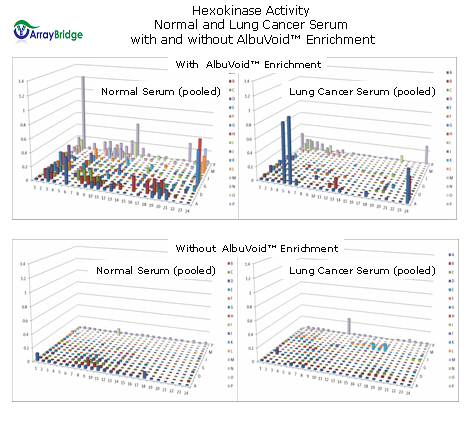
We report that by using AlbuVoid™ as an upfront serum enrichment product, the features displayed from the PEP technology is substantially improved compared to the virtually featureless landscape observed without any such enrichment. As an example using Hexokinase profiling, the final result is that with AlbuVoid™ enrichment, feature differences can be established between normal and cancer serum samples; such as would not be the case without AlbuVoid™ enrichment. Personal communication, Xing Wang, Ph.D., ArrayBridge (St. Louis, MO), manuscripts in process.
Knowledge surrounding the functional annotation of the proteome is vitally important as the landscape of protein conformations is highly variable, each conformation may contribute to a unique functional activity. Sequence annotation alone cannot capture this vital information, so new strategies are necessary. The challenge has been to overcome the analytical bias towards the most abundant proteins, and the complexities of mining the data to a manageable number of biomarker proteins that can be analyzed in more depth. So reconciling protein identifications to actual enzyme activities or functions has been subject to limitations in proteome separation and assay technologies. To overcome these inefficiencies in functional annotation, a top-down approach, starting with function, and ending with sequence and structural annotation is now available. The PEP technology, developed by ArrayBridge, uses a modified Two-dimensional Gel Electrophoresis to separate the proteome, without substantially compromising function. The isolated proteins are then electro-eluted from the PEP plate, and enzyme activities are measured systematically. This method thus provides a new functional dimension to explore the human serum proteome.
Genetic Engineering News Omics Tutorial Describes Biotech Support Group and ArrayBridge Joint Functional Proteomic Data
A Genetic Engineering News article describes a joint collaboration with ArrayBridge (St. Louis, MO). The article describes the combination of first low abundance protein enrichment/high abundance protein depletion with the Biotech Support Group product – AlbuVoid™, followed by a modified 2-dimensional electrophoretic separation, and transfer via the ArrayBridge PEP plate into microplates. From there, the resolved functionally active proteins were measured and characterized.
The citation is:
Xing Wang, Ph.D., Zhenyu Sun, M.D., Xiaofeng Chen, M.D., Xiong Su, Ph.D., Gan Wang, Ph.D., Matthew Kuruc. Genetic Engineering News Jan 1, 2015 (Vol. 35, No. 1) OMICS Tutorial Discovery of Functional Serum Biomarkers, Exploring Cancer's Signature in the Sensitive Functional Domain of the Human Proteome
http://www.genengnews.com/gen-articles/discovery-of-functional-serum-bio...
Serum Profiling Making Mark On Predictive Medicine
Vicki Glaser.Genetic Engineering & Biotechnology News. 2011;31(7):1-55.
AlbuVoid™'s application for albumin depletion is cited for inhibiting albumin binding and collecting the fraction containing albumin in Genetic Engineering & Biotechnology News.
Plasma
Espes, Daniel, Joey Lau, and Per-Ola Carlsson. "Increased circulating levels of betatrophin in individuals with long-standing type 1 diabetes." Diabetologia(2013): 1-4.
Authors Espes et al published an article in the journal Diabetologia which discovered an increase in hormone betatrophin in type 1 diabetics as compared to health individuals. Betatrophin causes an increase of pancreatic β cell replication and regulates glucose levels. Found in liver and adipose tissue, the hormone also increases β cell mass expansion. Analysis of betatrophin by mass spectrometry (MS) or gel electrophoresis could require enriching the protein mixture and depleting high abundance proteins to display less abundant proteins. The article titled, “Increased circulating levels of betatrophin in individuals with long-standing type 1 diabetes” utilized AlbuVoid™’s albumin depletion protocol for removing albumin from plasma samples. The article quotes AlbuVoid™ from Biotech Support Group, “Plasma samples were depleted of albumin using AlbuVoid Albumin Depletion Kit (Biotech Support Group, Monmouth Junction, NJ, USA). Data were normalized for total protein content.” Betatrophin increases proliferation of beta cell and this study identified double the concentration of betatrophin as measured by western immunoblot using a betatrophin primary antibody in type 1 diabetics. Moreover factors such as age in healthy controls displayed a direct relationship with increase in betatrophin whereas triacylglycerol, LDL-cholesterol, HDL-cholesterol levels and insulin were not affected. Increasing concentrations of betatrophin did not prevent against the loss of C-peptide suggesting type 1 diabetes on betatropin treatment would benefit from combination treatment. The authors concluded, “An intervention in patients with type 1 diabetes with betatrophin treatment might require supraphysiological dosing as well as combination with immune regulatory treatment.”
Cell Culture
Narain, Niven Rajin, Rangaprasad Sarangarajan, Vivek K. Vishnudas, and Michael Andrew Kiebish. "USE OF MARKERS IN THE IDENTIFICATION OF CARDIOTOXIC AGENTS AND IN THE DIAGNOSIS AND MONITORING OF CARDIOMYOPATHY AND CARDIOVASCULAR DISEASE." U.S. Patent 20,140,100,128, issued April 10, 2014
Narain, Niven Rajin et al authored a patent on methods for detecting biomarkers of cardiovascular diseases, monitoring disease progression and treatment of cardiovascular disease. Serum albumin is depleted from these samples using Biotech Support Group's AlbuVoid™ column.
The patent quotes "In one embodiment, the cells can be cultured in serum containing medium: The volume of the medium can be reduced using 3k MWCO Vivaspin columns (GE Healthcare Life Sciences), then can be reconstituted with 1×PBS (Invitrogen®). Serum albumin can be depleted from all samples using AlbuVoid column (Biotech Support Group, LLC) following the manufacturer's instructions with the modifications of buffer-exchange to optimize for condition medium application." Samples are labeled, vacuumed and analyzed by LC-MS/MS. The labeled peptide mixtures are separated by 2D-nanoLC and electrospray tandem mass spectrometry is used for analysis. Peptide mixtures are separated using a polysulfoethyl aspartamide column and eluted in a C18 trap column. Proteins and peptides are queried using bioinformatics software.
Patents
Patent Application Cites AlbuVoid™ For Secretome Sample Preparation & Neuroscience Research
Narain, Niven Rajin, and Paula Patricia Narain. COMPOSITIONS AND METHODS FOR DIAGNOSIS AND TREATMENT OF PERVASIVE DEVELOPMENTAL DISORDER United States Patent Application 20150023949, Pub. Date 01/22/2015.
The patent provides albumin sample preparation research application to track modulators of disease processes and experiment data on metabolomics, transcriptomics, single nucleotide polymorphisms using sample preparation and an artificial intelligence based informatics platform. Serum containing media are reduced using columns, reconstituted with buffers and serum albumin is depleted using Biotech Support Group's AlbuVoid™. Culture medium optimization and subsequent modifications to buffer exchange is performed. Protein extraction requires a lysis buffer with protease inhibitors, vortexing, ultrasonication, centrifugation and clean up of debris.
Narain, Niven Rajin, Rangaprasad Sarangarajan, and Vivek K. Vishnudas. "INTERROGATORY CELL-BASED ASSAYS AND USES THEREOF." U.S. Patent No. 20,120,258,874. 11 Oct. 2012.
Serum albumin depletion for secretome sample preparation using AlbuVoid™ while developing a cellular modeling system.
Inventors Narain et al published United States Patent 20120258874 titled, “INTERROGATORY CELL-BASED ASSAYS AND USES THEREOF” describing the invention of a cellular modeling system which develops molecular signatures allowing scientists to gain insight into the mechanisms of disease by providing information on tissue microenvironment. Researchers used AlbuVoid™ for serum albumin depletion for secretome sample preparation. Albumin was depleted from cell culture in serum containing medium using AlbuVoid™ to isolate and identify low abundance proteins.
The patient cites AlbuVoid™ from Biotech Support Group
“In one embodiment, the cells can be cultured in serum containing medium: The volume of the medium can be reduced using 3k MWCO Vivaspin columns (GE Healthcare Life Sciences), then can be reconstituted withlxPBS (Invitrogen). Serum albumin can be depleted from all samples using AlbuVoid column (Biotech Support Group, LLC) following the manufacturer's instructions with the modifications of buffer-exchange to optimize for condition medium application.”
Poster
Improved proteomic enrichment and workflow strategies
US HUPO 2014, Poster 089
Proteomic sample preparation technology for depleting hemoglobin, and enriching the low abundance proteome from human erythrocyte lysates and determination of the position and nature of human protein N termini in different tissues and disease states. Biotech Support Group has developed a series of enrichment reagents that offer common features and key advantages. After separations, the sub-proteomes retain their structural and functional integrity. All the products are consumable and economical; not being derived from biological sources. We report here applications for these products to specifically deplete albumin by a voidance strategy - called AlbuVoid™, which consequently enriches for the low abundance serum protein content. Such a strategy compares favorably to high abundance immuno-depletion (Agilent column) methods. Also, we continue to advance simplified workflows that utilize on-bead digestion for LC-MS. Together, these methods create efficiencies necessary to support high throughput investigations in all areas of discovery, targeted, functional and chemical proteomics.
New On-Bead Digestion Protocols Improve LC-MS Workflows Of Albumin Depleted Samples US HUPO
2013
Translational Proteomics
Biotech Support Group LLC, reports on a new technical poster which describes the use of on-bead digestion protocols as a suitable method to generate peptides for LC-MS/MS analysis, after albumin depletion on its AlbuVoid™ product. Entitled “New On-Bead Digestion Protocols Improve LC-MS Workflows Of Albumin Depleted Samples”, it was presented at the US HUPO Conference in Baltimore, MD, March 10-11, 2013. It describes by several performance metrics, that the ‘on-bead’ digestion protocol is comparable to the product’s standard elution protocol. On-bead protocols provide advantaged speed, simplicity and reduced potential for keratin contamination. The poster also reports on the efficiency of AlbuVoid™ as a simple consumable product for albumin depletion, with 20-30% more protein/peptide identifications achievable relative to untreated plasma, under identical operating and informatic parameters. “With greater interest in the proteomics community for better workflows and performance for LC-MS analyses, direct proteolytic digestion of the bound protein content - commonly called on-bead digestion, has many desirable workflow advantages. With this report on AlbuVoid™, we have now demonstrated that the surface chemistry platform from which it was derived, our novel passivated porous silica –NuGel™, offers the rigidity and low water content ideal for protein concentration and proteolytic digestion”, states Swapan Roy, Ph.D, President & Founder of Biotech Support Group. “We envision designing new strategies for high abundance depletion that can utilize on-bead digestion workflows for highly complex bead-bound proteomes along with many chemical proteomic applications.”
