Sample Prep - Pandemic Research
“… studies suggest that at least a subset of severe COVID-19 infection involves a catastrophic, complement-mediated thrombotic microvascular injury syndrome with sustained activation… of complement.” Translational Research 2020. Erythrocytes do not have a nucleus, and being in continuous contact with complement proteins in plasma, have a different make-up of complement regulators than nucleated cells. Increased complement activation or decreased complement regulation may result in red cell dysfunction. This may explain some of the clinical symptoms seen in severe COVID-19 patients- low oxygen levels and neurological complications. Therefore, proteomic investigation of erythrocytes is warranted. The hemoglobin removal products that follow have proven extremely efficient in coverage, detecting post-translational modifications, and quantifying the erythrocyte proteome. HemoVoid™Hemoglobin Depletion For Erythrocyte Proteomics

2DE Comparison. Key ReferencesThis study aimed to report the post-translational identity, biological activity and concentration of monomethyl and dimethylarginine-modified proteins by using GC-MS and LC-MS/MS approaches. The article states “we included in our method the use of HemoVoid™ … to improve the SDS-PAGE separation of proteins for further processing. HemoVoid™ …allowed removal of erythrocytic hemoglobin to a large extent. … removal of hemoglobin by this technique enabled an effective separation by SDS-PAGE and isolation of bands, presumably by avoiding overloading of the gels by hemoglobin.”. Bollenbach, Alexander, et al. "GC-MS and LC-MS/MS pilot studies on the guanidine (NG)-dimethylation in native, asymmetrically and symmetrically NG-dimethylated arginine-vasopressin peptides and proteins in human red blood cells." Journal of Chromatography B (2020): 122024. The article describes the advantage of HemoVoid™ in detection of low abundance proteins when comparing their amounts (in percent) between with four other extraction conditions. Most peptides, following HemoVoid™ extraction, “showed ion abundances ranging between 1.00E+5 and 1.00E+6 (31%). In comparison to this, fewer peptides (10–23%) were within this range following extraction with all other protocols”. With respect to potential biomarkers for Parkinson’s Disease, the article states “For example, PRDX6 accounts for 0.4% of the total ion abundance after DOC (deoxycholate) extraction, whereas following HV (HemoVoid™) extraction, this increases to 8%, a 20-fold enrichment”. The authors conclude that the HemoVoid™ method significantly reduces the concentration of hemoglobin, resulting in an increased signal-to noise of the remaining red cell proteins. The article describes methods to digest the HemoVoid™ bead-bound proteome, greatly simplifying the workflow for LC-MS/MS analysis. Klatt, Stephan, et al. " Optimizing red blood cell protein extraction for biomarker quantitation with mass spectrometry." Analytical and Bioanalytical Chemistry (2020): 1-14. HemogloBind™Suspension Reagent to remove Hemoglobin to assist the study of lower respiratory tract disease based on testing of bronchoalveolar lavage (BALF) fluid
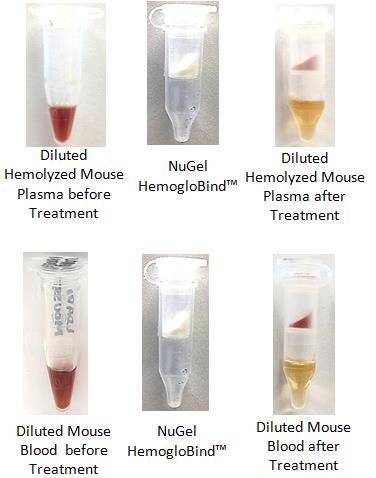
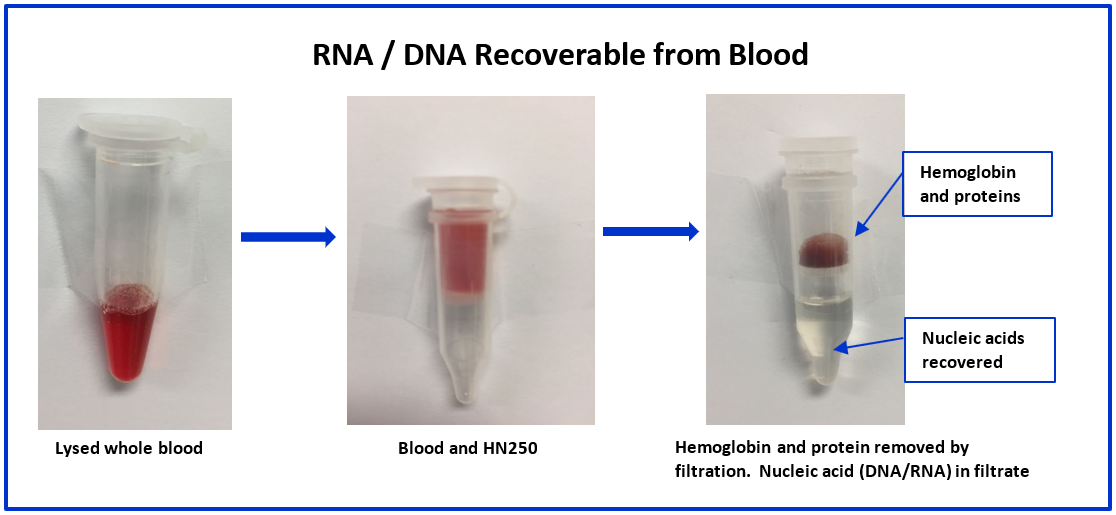
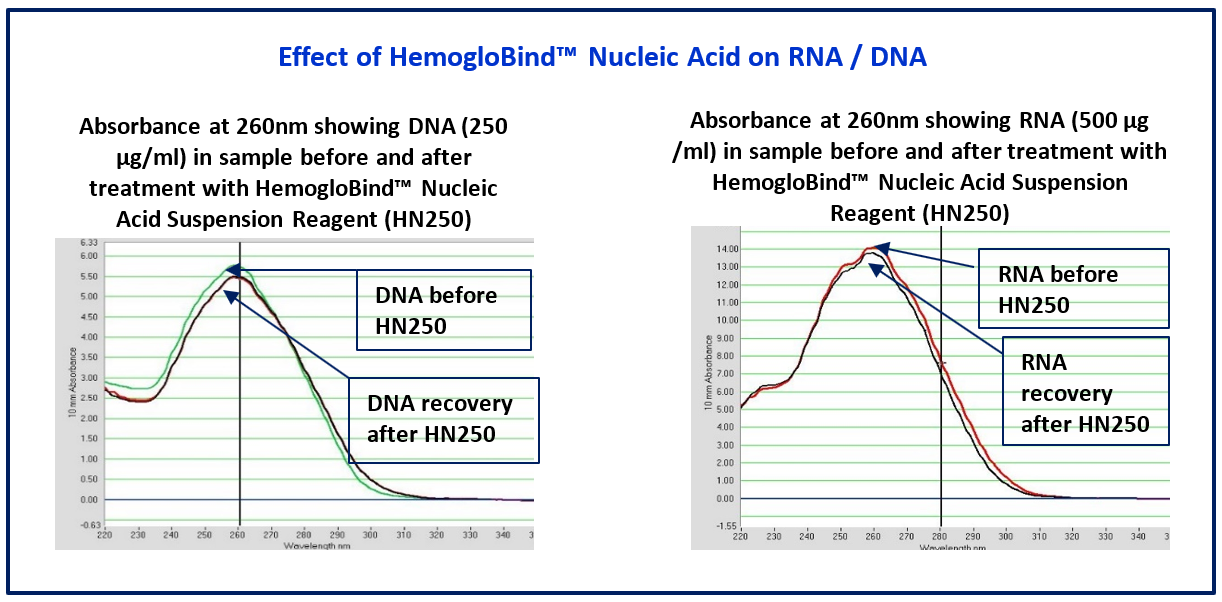
Key ReferencesHemogloBind ™ Cited in Comparison Proteomics Study of Covid-19 Patients in Peripheral Blood Derived Mononuclear Cells (PBMCs) – The Barometer of the Immune System HemogloBind ™ sample preparation technology provides deep and quantitative LC-MS proteome and phosphoproteome analysis. This study provides a comparison of SARS-CoV-2 positive ICU patients with age- and sex-matched SARS-CoV-2 negative ICU patients and healthy individuals, using peripheral blood derived mononuclear cells (PBMCs). Kaneko, Tomonori, et al. "System-wide hematopoietic and immune signaling aberrations in COVID-19 revealed by deep proteome and phosphoproteome analysis." Research Square preprint (2021). A research article describes the simplicity and efficiency of BSG's hemoglobin depletion technology for 2X improvement of proteomic information obtained from bronchoalveolar lavage fluid (BALF). Maneesh Bhargava M. et al."Proteome Profiling in Lung Injury after Hematopoietic Stem Cell Transplantation". Biol Blood Marrow Transplant 22 (2016) 1383-1390. http://www.bbmt.org/article/S1083-8791(16)30060-X/fulltext The cellular thermal shift assay (CETSA) enables proteome-wide target screening for unmodified antimalarial compounds with undetermined mechanisms of action, providing quantitative evidence about direct drug–protein interactions. The article states “The intact-cell CETSA protocol features a HemogloBind- based sample processing step, which provides a relatively fast, reliable and inexpensive method to deplete >90% of hemoglobin from processed intact-cell samples. As a result, it leads to a 40-50% increase in the number of peptide spectrum matches (PSMs) for P. falciparum and non-hemoglobin human proteins.”. The citation is: Dziekan, Jerzy Michal, et al. "Cellular thermal shift assay for the identification of drug–target interactions in the Plasmodium falciparum proteome." Nature Protocols (2020): 1-41. HemogloBind™ Nucleic AcidSuspension Reagent to remove Hemoglobin interference for PCR
Cleanascite™Lipid Removal Reagent and Clarification
Cleanascite™ is derived through a proprietary formulation of metallic oxide derivatives. Unlike other metallic oxides, Cleanascite™ does not have significant protein binding making its selectivity profile for lipids unique in the bio-research products industry. As a result, it is ideal to clear lipid-associated matrix effects from human sera, bile, ascites, and other high lipid content sample types. View Details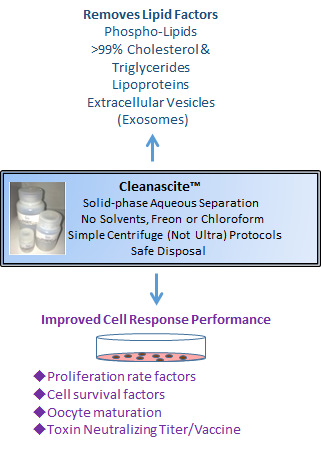

Cleanascite™ is supplied as a suspension reagent. Simply add, mix and centrifuge in a 10 minute protocol. Key ReferencesBiofluids
Farina, Annarita. "
Pre-fractionation of Noncirculating Biological Fluids to Improve Discovery of Clinically Relevant Protein Biomarkers." Proteomics for Biomarker Discovery. Humana Press, New York, NY, 2019. 23-37. Plasma/Serum Protein BiomarkersThe authors aimed at simultaneously measuring intact insulin and proinsulin derived C-peptide, to help predict development of diabetes mellitus, as well as in differential diagnosis in cases of hypoglycemia. Cleanascite™ is shown both to improve LC-MS measurements, and validated in accordance with CLIA ’88 guidelines. { doi: 10.1016/j.cca.2016.01.019 } Vaccine DevelopmentTo evaluate immunogenic response to a vaccine candidate, it is necessary to measure the antibodies from sera; a sample with a diverse lipid profile. In this citation, Cleanascite™ was used in a toxin neutralizing assay to evaluate the influence of cholesterol dependency, on a candidate protein pneumococcal vaccine. { doi: 10.1016/j.vaccine.2012.11.005 } Ascites Monoclonal AntibodiesThe researchers determined the role of complement on MAb-mediated protection for four mice Ig subclasses. After centrifugation of ascetic fluid, Cleanascite™ protocol was implemented to remove lipids. { doi: 10.1128/IAI.70.5.2598-2604.2002 } Cellular Response ApplicationsThe applications and references for the many diverse investigations using Cleanascite™ upstream of cell response measurements are described. { Cleanascite Cell Response Reference Applications } Tracheal Swab SamplesLi D, Wang J, Wang R, Li Y. A nanobeads amplified QCM immunosensor for the detection of avian influenza virus H5N1, Biosensors and Bioelectronics.2011;26(S10):4146-4154 Fu LM, Shinnick TM. Genome-wide exploration of the drug action of capreomycin on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips Journal of Infection.2007;54(S3):277-284 Fu LM, Shinnick TM. Genome-wide analysis of intergenic regions of mycobacterium tuberculosis H37Rv using affymetrix genechips. EURASIP journal on bioinformatics & systems biology.2007:23054 SalivaLucy E. DesJardin Isolation of M. tuberculosis RNA from Sputum Methods in Molecular Medicine.2001;48:133-139 Beenhouwer DO, Shapiro S, Feldmesser M et al. Both Th1 and Th2 Cytokines Affect the Ability of Monoclonal Antibodies To Protect Mice against Cryptococcus neoformans Infection and immunity.2001;69: 6445-6455 Desjardin LE, Perkins MD, Wolski K et al. Measurement of Sputum Mycobacterium tuberculosis Messenger RNA as a Surrogate for Response to Chemotherapy American journal of respiratory and critical care medicine.1999;160(1):203-10 Viraffinity™ & ViraPrep™ Mammal KitVirus Enrichment & Purification
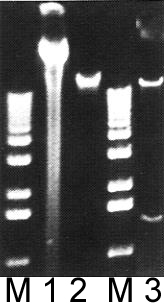
Lanes M: Markers
Since the COVID-19 virus can be detected in stool samples, it has been proposed that detection of the virus in wastewaters will help to monitor local or regional outbreaks. For this to succeed, simple and robust measurement of viruses from wastewater is necessary. Conventionally, enteric viruses are first concentrated by a variety of filter methods. A secondary enrichment step is often employed, to reduce to a final volume and improve purity prior to final analysis. This secondary step may be provided by the Viraffinity™ reagent, and does not require ultracentrifugation. As described in these two references: 1) Viraffinity™ can dramatically improve the purification of viruses, and 2) can remove RT-PCR inhibitors in the detection of enteric viruses. 1) Romain Fragnoud R., et al. "Differential proteomic analysis of virus enriched fractions obtained from plasma pools of patients with dengue fever or severe dengue". BMC Infectious Diseases (2015) 15:518. http://www.ncbi.nlm.nih.gov/pubmed/26572220 The article's authors report a method to compare the proteomes of virion-enriched fractions purified from plasma pools of patients with dengue fever or severe dengue. Virions were purified by ultracentrifugation combined with Viraffinity™. “…and became more intense after the Viraffinity™ step...Densitometry indicated that the protein complexity of the purified samples was reduced by roughly 350-fold compared to the unpurified samples.”. 2) Leggitt, Paris R., and Lee-ann Jaykus. "Detection methods for human enteric viruses in representative foods." Journal of Food Protection 63.12 (2000): 1738-1744. “To optimize viral nucleic acid amplification, secondary PEG precipitates… required a prior adsorption step with an equal volume of Viraffinity™ to further remove RT-PCR inhibitors. In this case, viruses …were adsorbed by the addition of an equal volume of Viraffinity™, … These Viraffinity™ precipitates were used directly in subsequent RNA extractions.” Viraffinity™Prep Size: Application dependent View DetailsViraPrep™ MammalPrep Size: 40 ml View DetailsA research article describes the simplicity and efficiency of this virus enrichment technology to enhance viral proteomic analyses from plasma. Romain Fragnoud R., et al. "Differential proteomic analysis of virus enriched fractions obtained from plasma pools of patients with dengue fever or severe dengue". BMC Infectious Diseases (2015) 15:518. http://www.ncbi.nlm.nih.gov/pubmed/26572220 The article's authors report a method to compare the proteomes of virion-enriched fractions purified from plasma pools of patients with dengue fever or severe dengue. Virions were purified by ultracentrifugation combined with Viraffinity™. “…and became more intense after the Viraffinity™ step...Densitometry indicated that the protein complexity of the purified samples was reduced by roughly 350-fold compared to the unpurified samples.". The authors conclude that the Viraffinity™ based technique of virion-enrichment allowed them to identify two host proteins that have prognostic value for classifying patients with acute dengue who are more likely to develop a severe dengue.
***Disclaimer*** Our BSG AdvantageConsumableCost-effective, not derived from biologicals
On-Bead DigestionEfficient workflows, quality LC-MS/MS data
Enrichment/DepletionDiverse strategies, species agnostic
Functional IntegrityMaintained throughout all separations
|

- About
- Products
- Hemoglobin Removal Kits
- Lipid Removal & Clarification
- Urine Protein & Low Abundance Enrichment
- Class Specific Enrichment
- Sample Prep Mass Spectrometry
- Functional & Chemical Proteomics
- Genomic Sample Prep
- Accessories
- Technical Resources
- References
- Publications & Reports
- FAQs
- Case Studies
- NRicher™ Bead Platform Provides Unique Sub-Proteome Biases And Fit For Purpose Opportunities for Targeted LC-MS Quantification
- BSG Products To Assist in Analyzing Macrophage Polarization
- Ectodomain Shedding and Enrichment of the Soluble Membrane Proteome
- Investigate out of the Venn Diagram box
- Methods to selectively deplete or purify Hemoglobin from Dried Blood Spots (DBS)
- The Utility of HemoVoid™ is Demonstrated in 3 Proteomic Investigations Identifying Potential Disease Specific Biomarkers
- The 4 common features of our sample prep products, known as the BSG Advantage, are highlighted in a selection of journal references.
- AlbuVoid™ Workflows Advance Cell Secretome Proteomics
- Lipid Removal for Phenotypic Cell Response in Cancer Research
- The Influence of Sample Prep Bias on LC-MS Targeted Peptide Quantification in Serum Proteomics
- Re-imagining proteomics for developing precision medicine biomarkers of the innate immune response in SARS-CoV-2
- Patent Application Describes New Proteomic Methods to Monitor Protease Inhibitor Function During Covid-19 Infections
- Efficient Hemoglobin Removal Advances Red Cell Proteomics Offering Many New Insights Into Inflammation and Infectious Disease
- The Potential for New Blood Biomarkers in the Management of COVID-19 Disease
- Establishing the Utility of HemoVoid™ and HemogloBind™ as Enrichment Tools for Proteomic Analysis of Red Cells and Whole Blood in Parkinson’s Disease
- Species Diversity Supported By BSG Products
- Poster Report Describes Loss of Functional Serpin Activity In Cancer Patient Blood
- AlbuVoid™️ PLUS & AlbuSorb™️ PLUS Evaluating Different Windows of Observation Solves The Many Challenges of Serum Proteomics
- Tackling the Challenges of Serum Proteomics
- Lipid Removal Sample Prep for Cell Response Applications
- Sample Prep for Proteomic Analysis of Saliva
- Biotech Support Group Featured in Book, "Functional Proteomics – Methods and Protocols"
- Sample Prep Liquid Biopsy Products Suitable for Proteomic Profiling of a Variety of Body Fluid Sample Types
- Albumin and High Abundance Depletion
- Using HemogloBind™ as a Hemoglobin Binding Reagent
- Diverse technologies available for researchers to selectively bind or enrich exosomes and extracellular vesicles.
- Stroma Liquid Biopsy™ Biomarkers Profile Pan-Cancer Dysregulation of the Serum Proteome
- Diverse Depletion and Enrichment Technologies Enhance Simplicity and Efficiency of Obtaining Quality Proteomic Information
- Use On-Bead Digestion to Improve Time Required for Serum Digestion
- Using AlbuVoid™ as a Serum Protein Enrichment Kit in Functional Proteomics
- Using Cleanascite™ as a Lipid Absorption and Clarification Reagent
- Using HemoVoid to Remove Hemoglobin Before Analysis
- Blog
- Contact
- Liquid Biopsy