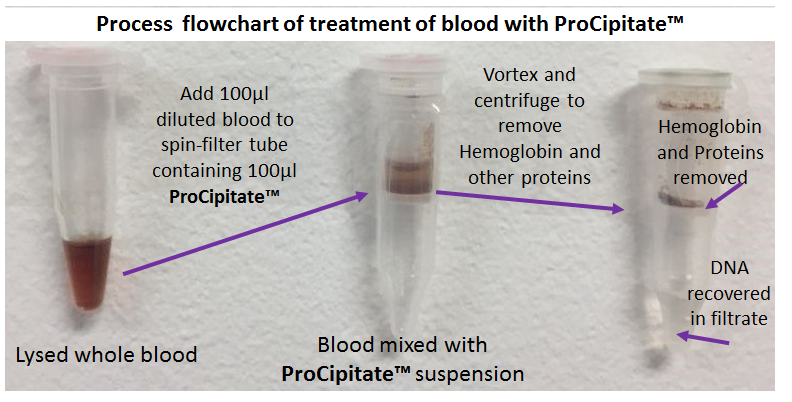
Genomic Sample PrepProCipitate™Superior Substitute to Phenol/Chloroform for Hemoglobin & Protein Removal, Isolation of DNA/RNA
PCR from whole blood using ProCipitate™ 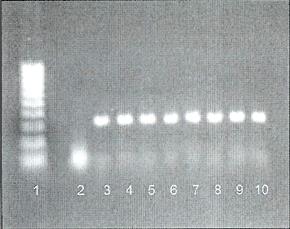
Lane 1: 100-1000 base pair Ladder
View Details Key References*J M Kelley et al. High Throughput Direct End Sequencing of BAC Clones. Nucleic Acids Research.1999.15;27(6):1539-1546 U.S. Patent Number 5,538,870, Method for Preparing Nucleic Acids For Analysis And Kits Useful Therefore. This patent shows the beneficial effects of ProCipitate™ in protocols which neutralize SDS with non-ionic detergents, are PCR compatible, and require no alcohol precipitation. ProCipitate™ appears in several articles and books on Food Safety Foodborne Disease Handbook, Second Edition,: Volume 2: Viruses: Parasites By Y. H. Hui, Sayed A. Sattar, Wai-Kit Nip “Viruses in the PEG eluants were precipitated…by an equal volume of ProCipitate™.” D’Souza, D. H. “Update on foodborne viruses: types, concentration and sampling methods.” Advances in Microbial Food Safety 2 (2014): 102. Health-related Water Microbiology, Volume 27, Issues 3-4, Pergamon, 1993 “ ProCipitate™ was an effective method to purify the sample and dramatically improve virus detectability by RT-PCR.” Cleanascite™Lipid Adsorption & Clarification
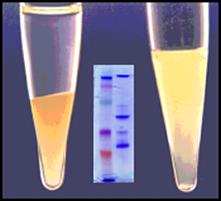
Viraffinity™ & ViraPrep™ KitsVirus Enrichment & Purification
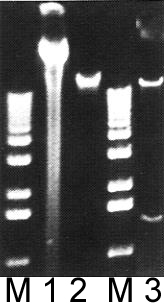
Lanes M: Markers
Viraffinity™Prep Size: Application dependent View DetailsViraPrep™ LambdaPrep Size: 5, 150mm platelysates View DetailsViraPrep™ MammalPrep Size: 40 ml View DetailsOur BSG AdvantageConsumableCost-effective, not derived from biologicals
On-Bead DigestionEfficient workflows, quality LC-MS/MS data
Enrichment/DepletionDiverse strategies, species agnostic
Functional IntegrityMaintained throughout all separations
|

- About
- Products
- Hemoglobin Removal Kits
- Lipid Removal & Clarification
- Urine Protein & Low Abundance Enrichment
- Class Specific Enrichment
- Sample Prep Mass Spectrometry
- Functional & Chemical Proteomics
- Genomic Sample Prep
- Accessories
- Technical Resources
- References
- Publications & Reports
- FAQs
- Case Studies
- Ectodomain Shedding and Enrichment of the Soluble Membrane Proteome
- Methods to selectively deplete or purify Hemoglobin from Dried Blood Spots (DBS)
- The Utility of HemoVoid™ is Demonstrated in 3 Proteomic Investigations Identifying Potential Disease Specific Biomarkers
- The 4 common features of our sample prep products, known as the BSG Advantage, are highlighted in a selection of journal references.
- AlbuVoid™ Workflows Advance Cell Secretome Proteomics
- Lipid Removal for Phenotypic Cell Response in Cancer Research
- The Influence of Sample Prep Bias on LC-MS Targeted Peptide Quantification in Serum Proteomics
- Re-imagining proteomics for developing precision medicine biomarkers of the innate immune response in SARS-CoV-2
- Patent Application Describes New Proteomic Methods to Monitor Protease Inhibitor Function During Covid-19 Infections
- Efficient Hemoglobin Removal Advances Red Cell Proteomics Offering Many New Insights Into Inflammation and Infectious Disease
- The Potential for New Blood Biomarkers in the Management of COVID-19 Disease
- Establishing the Utility of HemoVoid™ and HemogloBind™ as Enrichment Tools for Proteomic Analysis of Red Cells and Whole Blood in Parkinson’s Disease
- Species Diversity Supported By BSG Products
- Poster Report Describes Loss of Functional Serpin Activity In Cancer Patient Blood
- AlbuVoid™️ PLUS & AlbuSorb™️ PLUS Evaluating Different Windows of Observation Solves The Many Challenges of Serum Proteomics
- Tackling the Challenges of Serum Proteomics
- Lipid Removal Sample Prep for Cell Response Applications
- Sample Prep for Proteomic Analysis of Saliva
- Biotech Support Group Featured in Book, "Functional Proteomics – Methods and Protocols"
- Sample Prep Liquid Biopsy Products Suitable for Proteomic Profiling of a Variety of Body Fluid Sample Types
- Albumin and High Abundance Depletion
- Using HemogloBind™ as a Hemoglobin Binding Reagent
- Diverse technologies available for researchers to selectively bind or enrich exosomes and extracellular vesicles.
- Stroma Liquid Biopsy™ Biomarkers Profile Pan-Cancer Dysregulation of the Serum Proteome
- Diverse Depletion and Enrichment Technologies Enhance Simplicity and Efficiency of Obtaining Quality Proteomic Information
- Use On-Bead Digestion to Improve Time Required for Serum Digestion
- Using AlbuVoid™ as a Serum Protein Enrichment Kit in Functional Proteomics
- Using Cleanascite™ as a Lipid Absorption and Clarification Reagent
- Using HemoVoid to Remove Hemoglobin Before Analysis
- Contact
- Liquid Biopsy